

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145220.4 - phase: 0 /pseudo
(1391 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
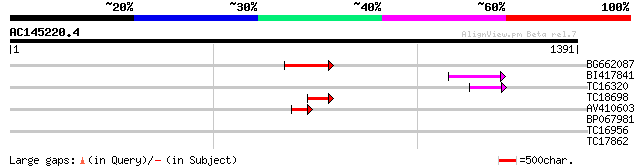
Score E
Sequences producing significant alignments: (bits) Value
BG662087 135 4e-32
BI417841 75 9e-14
TC16320 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial ... 61 1e-09
TC18698 57 2e-08
AV410603 51 1e-06
BP067981 29 5.8
TC16956 homologue to UP|Q9FJH0 (Q9FJH0) GTP-binding protein, ras... 28 7.6
TC17862 28 9.9
>BG662087
Length = 373
Score = 135 bits (340), Expect = 4e-32
Identities = 64/119 (53%), Positives = 84/119 (69%)
Frame = +1
Query: 675 GKVRMCVDFRDLNKASPKDNFPLPHIDMLVDNTAQPKVFSFMDGFSGYNQIKMSPEDREK 734
GK RM VD+ DLNKA PKD++PLP ID LVD + ++ S MD +SGY+QIKM P D +K
Sbjct: 16 GKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDEDK 195
Query: 735 TSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQH 793
T+F+T +CY+ +PFGL NAGATYQ M +F D + + +EVY+D+MIVKS H
Sbjct: 196 TAFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKSALRANH 372
>BI417841
Length = 617
Score = 74.7 bits (182), Expect = 9e-14
Identities = 43/142 (30%), Positives = 69/142 (48%), Gaps = 2/142 (1%)
Frame = +1
Query: 1076 GEGPDPNSKWGLVFDGAV--NAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACI 1133
G + N L FDG+ N G GAV+ + G + + + TNN AEY I
Sbjct: 109 GRQSNANRSCTLEFDGSSKGNPGSAGAGAVLRAEDGSKV-YLREGVGNQTNNQAEYRGLI 285
Query: 1134 FGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHI 1193
G++ A + +H+++ GDS LV Q++G W+ + N+ + A+ L + F +++H+
Sbjct: 286 LGLKHAHEQGYQHINVKGDSQLVCKQVEGSWKARNPNIASLCNEAKELKSKFQSFDINHV 465
Query: 1194 PRDENQMADALATLSSMFRVNH 1215
PR N AD A L H
Sbjct: 466 PRQYNSEADVQANLGVNLPAGH 531
>TC16320 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (11%)
Length = 632
Score = 60.8 bits (146), Expect = 1e-09
Identities = 30/91 (32%), Positives = 45/91 (48%)
Frame = +2
Query: 1129 YEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKV 1188
Y I G++ AI KH+ + GDS LV NQ++G W+ + N+ A+ L F
Sbjct: 2 YRGLILGLKHAIKEGYKHIQVKGDSMLVCNQVQGLWKIKNQNIASLCSEAKELKNKFLSF 181
Query: 1189 ELHHIPRDENQMADALATLSSMFRVNHWNDV 1219
+++HIPR+ N AD A R +V
Sbjct: 182 KINHIPREYNSEADVQANFGISLRAGQVEEV 274
>TC18698
Length = 808
Score = 57.4 bits (137), Expect = 2e-08
Identities = 31/62 (50%), Positives = 39/62 (62%)
Frame = -2
Query: 732 REKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEE 791
++KT+ + Y+VMP GL N TYQR M +FH I K VEVYV+DMIVKS+ E
Sbjct: 807 KKKTTLKINRVNYYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKSSQE* 628
Query: 792 QH 793
H
Sbjct: 627 FH 622
>AV410603
Length = 162
Score = 50.8 bits (120), Expect = 1e-06
Identities = 25/51 (49%), Positives = 33/51 (64%)
Frame = +1
Query: 692 KDNFPLPHIDMLVDNTAQPKVFSFMDGFSGYNQIKMSPEDREKTSFITPWG 742
KD+FP+P +D L+D + FS +D SGY+QI + PEDR KT F T G
Sbjct: 10 KDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRTHHG 162
>BP067981
Length = 334
Score = 28.9 bits (63), Expect = 5.8
Identities = 15/40 (37%), Positives = 27/40 (67%)
Frame = -2
Query: 65 SNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAH 104
S +D ++ ++ RSD+ +VD+ QT K +VL LLL++ +
Sbjct: 192 SAIDLVVVVVARSDFNVVDE-NQTEEKGTVLLLLLTTTTY 76
>TC16956 homologue to UP|Q9FJH0 (Q9FJH0) GTP-binding protein, ras-like
(At5g60860), partial (44%)
Length = 584
Score = 28.5 bits (62), Expect = 7.6
Identities = 20/80 (25%), Positives = 36/80 (45%), Gaps = 3/80 (3%)
Frame = -2
Query: 138 NLWFGEDELPEVGKCHNLALH---ISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRE 194
++WFG +ELP + H ++ S + + ++ G + P S++ L+ R
Sbjct: 343 DMWFGSEELPFYSLSNYAEQHPTFLTAETSSLLPTLIVCPLGKAAGSSPISSVFLLTTRY 164
Query: 195 IPLRRSTFLVKAFDGSRKNV 214
I +R S + F SR V
Sbjct: 163 I*VRTSVKALSTFKDSRAEV 104
>TC17862
Length = 545
Score = 28.1 bits (61), Expect = 9.9
Identities = 17/63 (26%), Positives = 29/63 (45%)
Frame = +2
Query: 962 ETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKNARWQMLLS 1021
E YTM + C L +++ +L+ H WL+ P ++I VT W+M S
Sbjct: 23 EDDYTMKRRLGCHL-----QVQIHLLRHLLWLLRHFLP*RHIRMMHLVTSCTLLWKM*CS 187
Query: 1022 EYD 1024
++
Sbjct: 188 SFE 196
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,155,564
Number of Sequences: 28460
Number of extensions: 346677
Number of successful extensions: 1634
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1571
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1633
length of query: 1391
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1289
effective length of database: 1,994,680
effective search space: 2571142520
effective search space used: 2571142520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC145220.4


