

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
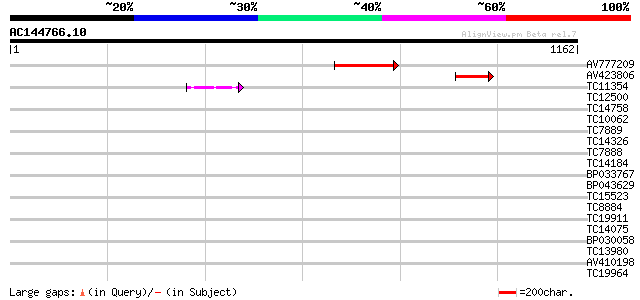
Query= AC144766.10 + phase: 2 /partial
(1162 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV777209 260 1e-69
AV423806 107 1e-23
TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II ... 42 7e-04
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 41 0.001
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 41 0.001
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 40 0.002
TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall prot... 40 0.003
TC14326 weakly similar to UP|EXLP_TOBAC (Q03211) Pistil-specific... 39 0.005
TC7888 similar to UP|Q40336 (Q40336) Proline-rich cell wall prot... 38 0.011
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 37 0.018
BP033767 37 0.018
BP043629 31 0.022
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 37 0.024
TC8884 similar to UP|O22600 (O22600) Glycine-rich protein, parti... 36 0.031
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 35 0.069
TC14075 homologue to UP|PRP2_MEDTR (Q40375) Repetitive proline-r... 35 0.069
BP030058 34 0.12
TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.3... 34 0.12
AV410198 34 0.12
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 34 0.15
>AV777209
Length = 393
Score = 260 bits (664), Expect = 1e-69
Identities = 119/130 (91%), Positives = 123/130 (94%)
Frame = +2
Query: 667 ETNRSEESPSHVHRLYFQGPNTFSEPWHLPHCPPEQVKDIVYEDAFNRFVDEINSLATYQ 726
ETNRSEES SHVHRLYFQGPNTFSEPWHLPH PPEQVKDIVYEDAFNRFVDE NSLATY
Sbjct: 2 ETNRSEESQSHVHRLYFQGPNTFSEPWHLPHYPPEQVKDIVYEDAFNRFVDEFNSLATYN 181
Query: 727 WWEGSIYTILCVTAYPLAWSWLQRCRRKKLQKLREFVRSEYDHACLRSCRSRALYEGLKV 786
WWEGSIY+ILC+ AYPLAWSWLQRCRR KLQKLREFVRSEYDHACLRSCRSRALYEG+KV
Sbjct: 182 WWEGSIYSILCIIAYPLAWSWLQRCRRMKLQKLREFVRSEYDHACLRSCRSRALYEGMKV 361
Query: 787 AATSDLMLAY 796
AAT+DLML Y
Sbjct: 362 AATTDLMLGY 391
>AV423806
Length = 233
Score = 107 bits (266), Expect = 1e-23
Identities = 52/77 (67%), Positives = 60/77 (77%)
Frame = +1
Query: 915 FGLVVHATENENMSSSGESYDDSRVTEKQSGFLRSPRNPVHHLTNNEQLLMPRRMSGGLL 974
FGLVVHATENE+ SSS E YDD R+ KQ LRSP NP H+T NEQLL+PRR+ GG+L
Sbjct: 1 FGLVVHATENESRSSSWEGYDDPRIAGKQPCLLRSPGNPACHMTGNEQLLLPRRIPGGIL 180
Query: 975 NGKILRTLKEKKTIYYP 991
+ K RTLKEKKTI+YP
Sbjct: 181 HPKSRRTLKEKKTIFYP 231
>TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II , partial
(6%)
Length = 583
Score = 41.6 bits (96), Expect = 7e-04
Identities = 40/123 (32%), Positives = 53/123 (42%), Gaps = 7/123 (5%)
Frame = +3
Query: 363 EGGTTYGDVDLPCELGSGSGNDSIAGATA----GGGIIVMGSLEHSLTSL-TLNGSLRSD 417
+GG G++D G G G + + G T GGG + + S L + GS+ D
Sbjct: 36 DGGVAVGELD-----GGGGGGEILGGETLEPELGGGELGDDDVGDSEAGLGRVEGSVFVD 200
Query: 418 GESFGDDIRRQDGRTSSIGPGGGSGGTVL--LFVQTLALGDSSIISTVGGQGSPSGGGGG 475
GE+ +D DG G G G GG V +A+GD G G GGGGG
Sbjct: 201 GEA--EDAVVVDGEGEDQGFGVGVGGWVFGEEVGGEVAVGD-------GDMGGDEGGGGG 353
Query: 476 GGG 478
G G
Sbjct: 354 GAG 362
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 41.2 bits (95), Expect = 0.001
Identities = 53/175 (30%), Positives = 66/175 (37%), Gaps = 13/175 (7%)
Frame = -2
Query: 318 SGVISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCEL 377
SGV+S +G GG G +GGG G G G G + E G D+ C
Sbjct: 584 SGVLSIEDIGRGGG---GMVATPYLGGGDGGCGGEGSGVWVIVVRELGGDGVVSDVRCVE 414
Query: 378 GSGSGNDSIAGATAGG--GIIVMGSLEHSLTSLTLNGSLRSDGESFGDDIRRQD------ 429
G GSG S+A + GG G++V G R G+ DI
Sbjct: 413 GGGSGALSVADMSKGGEGGMVVTPYFGGGGGGC--GGDGREQGKEASPDIIALPAITEAG 240
Query: 430 -----GRTSSIGPGGGSGGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGR 479
GR G GGG G L II+ +G+ GGGGGGGR
Sbjct: 239 GGVVGGRDGGGGCGGGGEGEQGRETSPTDLETFEIIALPAIKGAE--GGGGGGGR 81
Score = 39.7 bits (91), Expect = 0.003
Identities = 57/218 (26%), Positives = 76/218 (34%), Gaps = 34/218 (15%)
Frame = -2
Query: 341 GIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELGSGSGNDSIAGATAGGGIIVMGS 400
G+GGGGG G G G + + G + G L E G +A GGG G
Sbjct: 671 GVGGGGGGGDRVGVGGGASSDVSGVDSRGSGVLSIEDIGRGGGGMVATPYLGGGDGGCGG 492
Query: 401 LEHSLTSLTLNGSLRSDGESFGDDIRRQDGRTSSI---------------------GPGG 439
+ + + G+ D+R +G S G GG
Sbjct: 491 EGSGVWVIVVR---ELGGDGVVSDVRCVEGGGSGALSVADMSKGGEGGMVVTPYFGGGGG 321
Query: 440 GSGG------------TVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGRVHFHWSHI 487
G GG + L T A G VGG+ G GGGG G S
Sbjct: 320 GCGGDGREQGKEASPDIIALPAITEAGG-----GVVGGRDGGGGCGGGGEGEQGRETSPT 156
Query: 488 PVGD-EYITLASVEGSIITGGGFGGGQGLPGKNGSISG 524
+ E I L +++G+ GGG GGG+ L G G
Sbjct: 155 DLETFEIIALPAIKGA--EGGGGGGGRALSDFGGVEGG 48
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 40.8 bits (94), Expect = 0.001
Identities = 32/77 (41%), Positives = 34/77 (43%), Gaps = 7/77 (9%)
Frame = +1
Query: 325 GLGCTGGLGKGRYF------ENGIGGGGGHGGY-GGDGYYNGNFIEGGTTYGDVDLPCEL 377
G G GG G G Y E G GGGGG GY GG G Y G GG YG
Sbjct: 325 GGGGGGGRGGGGYGGGGGRREGGYGGGGGSRGYGGGGGGYGGG---GGGGYGGRREGGYG 495
Query: 378 GSGSGNDSIAGATAGGG 394
G G G+ +G GGG
Sbjct: 496 GDGGGSRYGSGGGGGGG 546
Score = 32.7 bits (73), Expect = 0.34
Identities = 42/149 (28%), Positives = 58/149 (38%), Gaps = 2/149 (1%)
Frame = +1
Query: 366 TTYGDVDLPCELGSGSGNDSIAGATAGGGIIVMGSLEHSLTSLTLNGSLRSDGESFG-DD 424
++YG++ + S ND G + G G + S E +++ DG + ++
Sbjct: 145 SSYGEI-----IDSKVVNDRETGRSRGFGFVTFTSEEAMRSAIEGMNGNELDGRNITVNE 309
Query: 425 IRRQDGRTSSIGPGGGS-GGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGRVHFH 483
+ + G G GGG GG G S GG G GGGGG GGR
Sbjct: 310 AQARGGGGGGGGRGGGGYGGGGGRREGGYGGGGGSRGYGGGGGGYGGGGGGGYGGRREGG 489
Query: 484 WSHIPVGDEYITLASVEGSIITGGGFGGG 512
+ G Y GS GGG GGG
Sbjct: 490 YGGDGGGSRY-------GS---GGGGGGG 546
Score = 31.6 bits (70), Expect = 0.76
Identities = 34/100 (34%), Positives = 36/100 (36%)
Frame = +1
Query: 344 GGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELGSGSGNDSIAGATAGGGIIVMGSLEH 403
GGGG GG G G Y G GG G G G G S GGG G +
Sbjct: 322 GGGGGGGGRGGGGYGGG---GGRREG--------GYGGGGGSRGYGGGGGGYGGGGGGGY 468
Query: 404 SLTSLTLNGSLRSDGESFGDDIRRQDGRTSSIGPGGGSGG 443
G R G +G DG S G GGG GG
Sbjct: 469 --------GGRREGG--YGG-----DGGGSRYGSGGGGGG 543
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 40.4 bits (93), Expect = 0.002
Identities = 29/84 (34%), Positives = 34/84 (39%)
Frame = +3
Query: 311 RSVKVEYSGVISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGD 370
R+ +E +G + G G G G GGG G GGYGG GY G + GG G
Sbjct: 219 RAKAIEVTGPNGQAVQGTRRGGGAG-------GGGRGGGGYGGGGYGGGGYGGGGRGGGG 377
Query: 371 VDLPCELGSGSGNDSIAGATAGGG 394
G G G G GGG
Sbjct: 378 ------YGGGGGYGGSGGYGGGGG 431
Score = 38.1 bits (87), Expect = 0.008
Identities = 21/41 (51%), Positives = 22/41 (53%)
Frame = +3
Query: 325 GLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGG 365
G G GG G+G G GG GG GGYGG G Y G GG
Sbjct: 336 GGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGGYGGGGRGGG 458
Score = 33.5 bits (75), Expect = 0.20
Identities = 28/86 (32%), Positives = 33/86 (37%), Gaps = 5/86 (5%)
Frame = +3
Query: 440 GSGGTVLLFVQTLALGDSSIISTVGG-----QGSPSGGGGGGGGRVHFHWSHIPVGDEYI 494
G G V V + G + I G QG+ GGG GGGGR + G
Sbjct: 171 GEGEDVEFIVASDPDGRAKAIEVTGPNGQAVQGTRRGGGAGGGGRGGGGYGGGGYGG--- 341
Query: 495 TLASVEGSIITGGGFGGGQGLPGKNG 520
G GGG+GGG G G G
Sbjct: 342 --GGYGGGGRGGGGYGGGGGYGGSGG 413
Score = 29.6 bits (65), Expect = 2.9
Identities = 30/96 (31%), Positives = 36/96 (37%)
Frame = +3
Query: 429 DGRTSSIGPGGGSGGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGRVHFHWSHIP 488
DGR +I G +G V + G G G GGG GGGGR +
Sbjct: 213 DGRAKAIEVTGPNGQAVQGTRRGGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGG-- 386
Query: 489 VGDEYITLASVEGSIITGGGFGGGQGLPGKNGSISG 524
G Y G GGG+GGG G+ G G
Sbjct: 387 -GGGY----GGSGGYGGGGGYGGGGRGGGRGGGGGG 479
Score = 28.9 bits (63), Expect = 4.9
Identities = 17/35 (48%), Positives = 18/35 (50%)
Frame = +3
Query: 325 GLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNG 359
G G GG G G G GGGGG+GG G G G
Sbjct: 369 GGGYGGGGGYGG--SGGYGGGGGYGGGGRGGGRGG 467
Score = 28.5 bits (62), Expect = 6.4
Identities = 15/35 (42%), Positives = 17/35 (47%)
Frame = +3
Query: 319 GVISASGLGCTGGLGKGRYFENGIGGGGGHGGYGG 353
G G G +GG G G + G GGG GG GG
Sbjct: 375 GYGGGGGYGGSGGYGGGGGYGGGGRGGGRGGGGGG 479
>TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (66%)
Length = 1158
Score = 39.7 bits (91), Expect = 0.003
Identities = 31/79 (39%), Positives = 36/79 (45%), Gaps = 6/79 (7%)
Frame = -2
Query: 321 ISASGLG--CTGGLGKGRYFENGIGGG----GGHGGYGGDGYYNGNFIEGGTTYGDVDLP 374
+S GLG TGG+G G G+G G GG G GG G G GG T G V
Sbjct: 479 VSVGGLGGVTTGGVGVGGVTIGGVGVGGVTIGGDGTTGGSGT*GGG*TIGGFTIGGV--- 309
Query: 375 CELGSGSGNDSIAGATAGG 393
G+ G +I G T GG
Sbjct: 308 ---GT*GGG*TIGGFTIGG 261
Score = 35.4 bits (80), Expect = 0.053
Identities = 28/75 (37%), Positives = 32/75 (42%), Gaps = 2/75 (2%)
Frame = -2
Query: 321 ISASGLGCTGGLGKGRYFENGIGGGGGHG-GYGG-DGYYNGNFIEGGTTYGDVDLPCELG 378
+S +GC GG G G GGGGGHG GG G G GG T G V +
Sbjct: 548 VSIGHVGCAGG-GDGT-----TGGGGGHGVSVGGLGGVTTGGVGVGGVTIGGVGVGGVTI 387
Query: 379 SGSGNDSIAGATAGG 393
G G +G GG
Sbjct: 386 GGDGTTGGSGT*GGG 342
Score = 29.3 bits (64), Expect = 3.8
Identities = 23/69 (33%), Positives = 27/69 (38%), Gaps = 4/69 (5%)
Frame = -2
Query: 460 ISTVGGQGSPSGGGGGG----GGRVHFHWSHIPVGDEYITLASVEGSIITGGGFGGGQGL 515
+ GG +GGGGG GG + VG I V G I G G GG G
Sbjct: 533 VGCAGGGDGTTGGGGGHGVSVGGLGGVTTGGVGVGGVTIGGVGVGGVTIGGDGTTGGSGT 354
Query: 516 PGKNGSISG 524
G +I G
Sbjct: 353 *GGG*TIGG 327
>TC14326 weakly similar to UP|EXLP_TOBAC (Q03211) Pistil-specific
extensin-like protein precursor (PELP), partial (11%)
Length = 630
Score = 38.9 bits (89), Expect = 0.005
Identities = 56/203 (27%), Positives = 70/203 (33%), Gaps = 8/203 (3%)
Frame = -1
Query: 321 ISASGLGCTGGLGKGRYFENGIGGG----GGHGGYGGDGYYNGNFIEGGT---TYGDVDL 373
+ G+G TGG G G GGG GG G + G G G+F GG T GD +
Sbjct: 576 MGVDGIGATGGFTTG-----G*GGGVFTWGGVGAFTGGG**GGDFTMGGVGAFTGGDFTM 412
Query: 374 PCELGSGSGNDSIAGATAGGGIIVMGSLEHSLTSLTLNGSLRSDGESFGDDIRRQDGRTS 433
G + G GG MG + G G D G
Sbjct: 411 -------GGEGAFTGGG*EGGDFTMGGV----------------GAFTGGDFTM--GGEG 307
Query: 434 SIGPGGGSGGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGRVHFHWSHIPVGDEY 493
+ GG GG T+GG+G+ +GG GGG G G
Sbjct: 306 AFTGGGK*GGDF----------------TMGGEGAFTGGCGGGVGA-------FTRGGVG 196
Query: 494 ITLASVEG-SIITGGGFGGGQGL 515
A VEG TG G G +G+
Sbjct: 195 TVTAGVEGVGGFTGEGGAGTEGI 127
Score = 38.5 bits (88), Expect = 0.006
Identities = 46/156 (29%), Positives = 65/156 (41%)
Frame = -1
Query: 321 ISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELGSG 380
I A+G TGG G G + G+G G G GGD G G T GD + E G+
Sbjct: 561 IGATGGFTTGG*GGGVFTWGGVGAFTGGG**GGDFTMGG---VGAFTGGDFTMGGE-GAF 394
Query: 381 SGNDSIAGATAGGGIIVMGSLEHSLTSLTLNGSLRSDGESFGDDIRRQDGRTSSIGPGGG 440
+G G GG+ G+ ++ G+ G+ GD +G + GG
Sbjct: 393 TGGG*EGGDFTMGGV---GAFTGGDFTMGGEGAFTGGGK*GGDFTMGGEGAFT-----GG 238
Query: 441 SGGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGG 476
GG V F + G ++ + V G G +G GG G
Sbjct: 237 CGGGVGAFTRG---GVGTVTAGVEGVGGFTGEGGAG 139
>TC7888 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (60%)
Length = 913
Score = 37.7 bits (86), Expect = 0.011
Identities = 65/226 (28%), Positives = 79/226 (34%), Gaps = 29/226 (12%)
Frame = -1
Query: 325 GLG--CTGGLGKGRYFENGIGGG----GGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELG 378
GLG TGG+G G G+G G GG G GG G G GG T G V G
Sbjct: 913 GLGGVTTGGVGVGGVTIGGVGVGGVTIGGDGTTGGSGT*GGG*TIGGFTIGGV------G 752
Query: 379 SGSGNDSIAGATAG----------------GGIIVMGSLEHSLTSLTLNGSLRSDGESFG 422
+ G +I G T G GG G + + L G + FG
Sbjct: 751 T*GGG*TIGGFTIGGFGT*GGG*TMGGFTMGGFGT*GGVTGTTGGLGTYGGVTGTTGGFG 572
Query: 423 DDIRRQDGRTSSIGPGGGSGGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGRV-- 480
G + G G +GG LG T+GG G+ G G GG +
Sbjct: 571 T*GGETTGGLGT*GGGFTTGG----------LGT*GGF-TIGGLGTWGGFGT*GGFTIGG 425
Query: 481 HFHWSHIPVGDEYITLASVEGSIITG-----GGFGGGQGLPGKNGS 521
W G G + TG GGFG G G G +GS
Sbjct: 424 FGTWGGFTTGG-----FGT*GGLTTGGLG**GGFGDGLG--GGHGS 308
Score = 35.4 bits (80), Expect = 0.053
Identities = 32/89 (35%), Positives = 37/89 (40%), Gaps = 5/89 (5%)
Frame = -1
Query: 319 GVISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELG 378
G ++ GLG GG G G GGGHG DG Y G GG T G C G
Sbjct: 379 GGLTTGGLG**GGFGDGL--------GGGHGSGP*DGGYFGGL*TGGFTIGG----CCGG 236
Query: 379 SGSGNDSIAGATAG---GGIIV--MGSLE 402
+G +I G G GG + G LE
Sbjct: 235 L*TGGFTIGGCCGGL*TGGFTIGCFGGLE 149
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 37.0 bits (84), Expect = 0.018
Identities = 25/65 (38%), Positives = 29/65 (44%)
Frame = +3
Query: 322 SASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELGSGS 381
S G G TGG G G G G GGG GG G G + GG YG+ D SG+
Sbjct: 363 SGGGYG-TGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDD------SGN 521
Query: 382 GNDSI 386
D +
Sbjct: 522 RRDEV 536
Score = 31.6 bits (70), Expect = 0.76
Identities = 36/129 (27%), Positives = 42/129 (31%), Gaps = 6/129 (4%)
Frame = +3
Query: 411 NGSLRSDGESFGDDIRRQDGRTSSIG---PGGGSGGTVLLFVQTLALGDSSIISTVGGQG 467
+G +S E+ G +TSS P G GG DS T
Sbjct: 210 SGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGY-----------DSGYNKTAYSTD 356
Query: 468 SPSGGGGGGGGRVHFHWSHIPVGDEYITLASVEGSIITGGGF---GGGQGLPGKNGSISG 524
PSGGG G GG G TGGG+ GGG G G G
Sbjct: 357 EPSGGGYGTGG--------------------AGGGYSTGGGYGTGGGGGGEYSTGGGGGG 476
Query: 525 KACPKGLYG 533
+ G YG
Sbjct: 477 YSTGGGGYG 503
Score = 31.6 bits (70), Expect = 0.76
Identities = 19/38 (50%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Frame = +3
Query: 319 GVISASGLGCTGGLGKGRYFENGIGGG--GGHGGYGGD 354
G + G G TGG G G Y G GGG G GGYG D
Sbjct: 399 GYSTGGGYG-TGGGGGGEYSTGGGGGGYSTGGGGYGND 509
>BP033767
Length = 541
Score = 37.0 bits (84), Expect = 0.018
Identities = 37/124 (29%), Positives = 52/124 (41%)
Frame = -3
Query: 389 ATAGGGIIVMGSLEHSLTSLTLNGSLRSDGESFGDDIRRQDGRTSSIGPGGGSGGTVLLF 448
A G +++G + + S G R DG+ GD +G + G G GG
Sbjct: 539 AAVGSESLLVGGVVFAAASAVDEGGDRDDGDGAGDG---GNGDAEELVGGFGFGG----- 384
Query: 449 VQTLALGDSSIISTVGGQGSPSGGGGGGGGRVHFHWSHIPVGDEYITLASVEGSIITGGG 508
++ GD VGG+G G GGGR + W + G+E G I+ GGG
Sbjct: 383 LREDEGGD------VGGEG------GEGGGRRRWWWRWV-CGEERC------GGILGGGG 261
Query: 509 FGGG 512
GGG
Sbjct: 260 CGGG 249
Score = 28.1 bits (61), Expect = 8.4
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 9/70 (12%)
Frame = -1
Query: 464 GGQGSPSGGGGGGGGR------VHFHWSHIPVGDE---YITLASVEGSIITGGGFGGGQG 514
G + G GGGGG V W + VG E Y EG GGGFGG +G
Sbjct: 352 GKEAKEEGEGGGGGDGYVGKKGVEAFWVVVVVGVEENGYFGFGG-EGDGDGGGGFGG*EG 176
Query: 515 LPGKNGSISG 524
G+ + G
Sbjct: 175 --GREPCVKG 152
>BP043629
Length = 562
Score = 31.2 bits (69), Expect(2) = 0.022
Identities = 26/78 (33%), Positives = 35/78 (44%), Gaps = 4/78 (5%)
Frame = -1
Query: 404 SLTSLTLNGSLRSDGESFGDD-IRRQDGRTSSIGPGGGSGGTVLLFVQTLALGDSSIIST 462
S T+L SL + E+F +D + + RT S G GG + L T G +S+
Sbjct: 508 STTALLSRDSLGASLENFHEDLVPSLNSRTGSAGGGGFA-----LVTTTFGSGCASLAGD 344
Query: 463 V---GGQGSPSGGGGGGG 477
G SGG GGGG
Sbjct: 343 TPCWSASGEESGGVGGGG 290
Score = 24.3 bits (51), Expect(2) = 0.022
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = -3
Query: 340 NGIGGGGGHGGYGGDGYYNGNFIEG 364
NG GGG G G G ++G+ IEG
Sbjct: 557 NGGGGGVGSGVKAPLGIHHGSAIEG 483
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 36.6 bits (83), Expect = 0.024
Identities = 28/69 (40%), Positives = 31/69 (44%), Gaps = 10/69 (14%)
Frame = +2
Query: 311 RSVKVEYSGVISASGLGCTGGLGKGRYFENGIGGGGG----------HGGYGGDGYYNGN 360
R+V V + G G GG G GR E G GGGGG +GG GG G Y G
Sbjct: 563 RNVTVNEAQARGRGGGGGGGGYGGGRR-EGGYGGGGGSRYSGGGGGGYGGSGGGG-YGGR 736
Query: 361 FIEGGTTYG 369
GG YG
Sbjct: 737 REGGGGGYG 763
Score = 32.0 bits (71), Expect = 0.58
Identities = 19/42 (45%), Positives = 21/42 (49%), Gaps = 4/42 (9%)
Frame = +2
Query: 322 SASGLGCTGGLGKGRYFENGIGGGGGHGGY----GGDGYYNG 359
S G G GG G G Y GGGGG+GG GG Y +G
Sbjct: 680 SGGGGGGYGGSGGGGYGGRREGGGGGYGGSREGGGGSRYSSG 805
Score = 31.6 bits (70), Expect = 0.76
Identities = 28/97 (28%), Positives = 36/97 (36%)
Frame = +2
Query: 383 NDSIAGATAGGGIIVMGSLEHSLTSLTLNGSLRSDGESFGDDIRRQDGRTSSIGPGGGSG 442
ND G + G G + S + ++ DG + + + GR G GG G
Sbjct: 455 NDRETGRSRGFGFVTFASEQSMKDAIEGMNGQDIDGRNVTVNEAQARGRGGGGGGGGYGG 634
Query: 443 GTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGGGR 479
G G S S GG G GGGG GGR
Sbjct: 635 GRR---EGGYGGGGGSRYSGGGGGGYGGSGGGGYGGR 736
Score = 28.9 bits (63), Expect = 4.9
Identities = 24/69 (34%), Positives = 28/69 (39%)
Frame = +2
Query: 465 GQGSPSGGGGGGGGRVHFHWSHIPVGDEYITLASVEGSIITGGGFGGGQGLPGKNGSISG 524
G+G GGGG GGGR + GS +GGG GGG G G G
Sbjct: 596 GRGGGGGGGGYGGGRREGGY------------GGGGGSRYSGGG-GGGYGGSGGGGYGGR 736
Query: 525 KACPKGLYG 533
+ G YG
Sbjct: 737 REGGGGGYG 763
>TC8884 similar to UP|O22600 (O22600) Glycine-rich protein, partial (46%)
Length = 888
Score = 36.2 bits (82), Expect = 0.031
Identities = 27/67 (40%), Positives = 32/67 (47%), Gaps = 7/67 (10%)
Frame = +1
Query: 315 VEYSGVISASGLGCTG------GLGKGRYFENGIGGG-GGHGGYGGDGYYNGNFIEGGTT 367
+ Y GV SG+G G G G G F+NG+GGG GG GG G G I GT
Sbjct: 445 MSYGGVGGYSGIGSNGLPFGTVGAGVGGGFDNGLGGGIGGTVSIGGPG--GGAGIGAGTG 618
Query: 368 YGDVDLP 374
G + P
Sbjct: 619 GGVLPHP 639
Score = 28.1 bits (61), Expect = 8.4
Identities = 25/76 (32%), Positives = 34/76 (43%)
Frame = +1
Query: 321 ISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCELGSG 380
+S G+G G+G NG+ G G GG G+ NG GG G V + G
Sbjct: 445 MSYGGVGGYSGIG-----SNGLPFGTVGAGVGG-GFDNG---LGGGIGGTVSI-----GG 582
Query: 381 SGNDSIAGATAGGGII 396
G + GA GGG++
Sbjct: 583 PGGGAGIGAGTGGGVL 630
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 35.0 bits (79), Expect = 0.069
Identities = 27/71 (38%), Positives = 35/71 (49%)
Frame = +1
Query: 318 SGVISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPCEL 377
SG + G G GG G Y ++G GGG GGYG G +G+ GG YGD
Sbjct: 157 SGGRTGGGYG-DGGHSAGGYGDHGGRSGGGGGGYGDGGLSSGS---GG--YGDGG---RS 309
Query: 378 GSGSGNDSIAG 388
G G G+ ++G
Sbjct: 310 GVGYGDGXLSG 342
Score = 29.6 bits (65), Expect = 2.9
Identities = 35/124 (28%), Positives = 44/124 (35%), Gaps = 10/124 (8%)
Frame = +1
Query: 411 NGSLRSDGESFG------DDIRRQDGRTSS-IGPGGGSGGTVLLFVQTLALGDSSIISTV 463
+G RS+ G DD R GRT G GG S G
Sbjct: 88 HGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYGDGGHSAGGY------------------ 213
Query: 464 GGQGSPSGGGGGGGGRVHFHWSHIPVGDEYITLASVEGSIITG-GGFGGG--QGLPGKNG 520
G G SGGGGGG G +G + +G GG+G G G+ +G
Sbjct: 214 GDHGGRSGGGGGGYG---------------------DGGLSSGSGGYGDGGRSGVGYGDG 330
Query: 521 SISG 524
+SG
Sbjct: 331 XLSG 342
>TC14075 homologue to UP|PRP2_MEDTR (Q40375) Repetitive proline-rich cell
wall protein 2 precursor, partial (98%)
Length = 1524
Score = 35.0 bits (79), Expect = 0.069
Identities = 27/78 (34%), Positives = 32/78 (40%), Gaps = 1/78 (1%)
Frame = -2
Query: 317 YSGVISASGLGCTGGLGKGRYFENGIGGGGGH-GGYGGDGYYNGNFIEGGTTYGDVDLPC 375
Y+G S GL TGG G + G GG + GG+ G Y G F GG G
Sbjct: 593 YTGGFSTGGL*-TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGFSTG- 420
Query: 376 ELGSGSGNDSIAGATAGG 393
G +G S G GG
Sbjct: 419 --GL*TGGFSTGGLYTGG 372
Score = 34.7 bits (78), Expect = 0.090
Identities = 27/78 (34%), Positives = 31/78 (39%), Gaps = 1/78 (1%)
Frame = -2
Query: 317 YSGVISASGLGCTGGLGKGRYFENGIGGGGG-HGGYGGDGYYNGNFIEGGTTYGDVDLPC 375
Y+G S GL TGG G + G GG GG+ G Y G F GG G
Sbjct: 383 YTGGFSTGGL*-TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTG- 210
Query: 376 ELGSGSGNDSIAGATAGG 393
G +G S G GG
Sbjct: 209 --GLYTGGFSTGGLYTGG 162
Score = 33.9 bits (76), Expect = 0.15
Identities = 26/77 (33%), Positives = 32/77 (40%), Gaps = 1/77 (1%)
Frame = -2
Query: 318 SGVISASGLGCTGGLGKGRYFENGIGGGGGH-GGYGGDGYYNGNFIEGGTTYGDVDLPCE 376
+G S GL TGG G + G GG + GG+ G Y G F GG G
Sbjct: 290 TGGFSTGGL-YTGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TG------- 135
Query: 377 LGSGSGNDSIAGATAGG 393
G +G I G +GG
Sbjct: 134 -GFSTGGL*IGGVYSGG 87
Score = 32.7 bits (73), Expect = 0.34
Identities = 26/77 (33%), Positives = 31/77 (39%), Gaps = 1/77 (1%)
Frame = -2
Query: 318 SGVISASGLGCTGGLGKGRYFENGIGGGGG-HGGYGGDGYYNGNFIEGGTTYGDVDLPCE 376
+G SA GL TGG G + G GG GG+ G Y G F GG G
Sbjct: 650 TGGFSAGGL*-TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTG------- 495
Query: 377 LGSGSGNDSIAGATAGG 393
G +G G + GG
Sbjct: 494 -GFSTGGLYTGGFSTGG 447
Score = 32.0 bits (71), Expect = 0.58
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 1/56 (1%)
Frame = -2
Query: 317 YSGVISASGLGCTGGLGKGRYFENGIGGGGGH-GGYGGDGYYNGNFIEGGTTYGDV 371
Y+G S GL TGG G + G GG + GG+ G G F GG G V
Sbjct: 263 YTGGFSTGGL-YTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGFSTGGL*IGGV 99
Score = 32.0 bits (71), Expect = 0.58
Identities = 21/54 (38%), Positives = 24/54 (43%), Gaps = 1/54 (1%)
Frame = -2
Query: 317 YSGVISASGLGCTGGLGKGRYFENGIGGGGGH-GGYGGDGYYNGNFIEGGTTYG 369
Y+G S GL TGG G G GG + GG+ G Y G F GG G
Sbjct: 833 YTGGFSTGGL*-TGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTG 675
Score = 31.2 bits (69), Expect = 0.99
Identities = 26/77 (33%), Positives = 30/77 (38%), Gaps = 1/77 (1%)
Frame = -2
Query: 318 SGVISASGLGCTGGLGKGRYFENGIGGGGG-HGGYGGDGYYNGNFIEGGTTYGDVDLPCE 376
+G S GL TGG G + G GG GG+ G Y G F GG G
Sbjct: 440 TGGFSTGGL*-TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTG-- 270
Query: 377 LGSGSGNDSIAGATAGG 393
G +G S G GG
Sbjct: 269 -GLYTGGFSTGGLYTGG 222
Score = 30.8 bits (68), Expect = 1.3
Identities = 26/78 (33%), Positives = 31/78 (39%), Gaps = 1/78 (1%)
Frame = -2
Query: 317 YSGVISASGLGCTGGLGKGRYFENGI-GGGGGHGGYGGDGYYNGNFIEGGTTYGDVDLPC 375
Y+G S GL TGG G + G+ GG GG+ G G F GG G
Sbjct: 743 YTGGFSTGGL-YTGGFSTGGLYTGGL*TGGL*TGGFSAGGL*TGGFSTGGLYTGGFSTG- 570
Query: 376 ELGSGSGNDSIAGATAGG 393
G +G S G GG
Sbjct: 569 --GL*TGGFSTGGLYTGG 522
Score = 29.3 bits (64), Expect = 3.8
Identities = 25/77 (32%), Positives = 30/77 (38%), Gaps = 1/77 (1%)
Frame = -2
Query: 318 SGVISASGLGCTGGLGKGRYFENGIGGGGGH-GGYGGDGYYNGNFIEGGTTYGDVDLPCE 376
+G S GL TGG G + G GG + GG+ G Y G GG G
Sbjct: 800 TGGFSTGGL*-TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGL*TGGL*TGGFSAG-- 630
Query: 377 LGSGSGNDSIAGATAGG 393
G +G S G GG
Sbjct: 629 -GL*TGGFSTGGLYTGG 582
Score = 29.3 bits (64), Expect = 3.8
Identities = 27/78 (34%), Positives = 31/78 (39%), Gaps = 1/78 (1%)
Frame = -2
Query: 317 YSGVISASGLGCTGGLGKGRYFENGIGGGGG-HGGYGGDGYYNGNFIEGGTTYGDVDLPC 375
Y+G + GL TGGL G G GG GG+ G Y G F GG G
Sbjct: 953 YTGGL*TGGL-YTGGL*TGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTG- 780
Query: 376 ELGSGSGNDSIAGATAGG 393
G +G S G GG
Sbjct: 779 --GL*TGGFSTGGLYTGG 732
>BP030058
Length = 455
Score = 34.3 bits (77), Expect = 0.12
Identities = 30/81 (37%), Positives = 36/81 (44%), Gaps = 18/81 (22%)
Frame = +1
Query: 325 GLGCTG-----------GL---GKGRYF----ENGIGGGGGHGGYGGDGYYNGNFIEGGT 366
GLGC G GL G+ YF E G GGGG GG GGDG+ +G GG
Sbjct: 214 GLGCPGTYMILGGGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGDGHGSG----GGG 381
Query: 367 TYGDVDLPCELGSGSGNDSIA 387
GD G G G+ ++
Sbjct: 382 GEGDG------GGGGGSPQVS 426
Score = 31.2 bits (69), Expect = 0.99
Identities = 22/67 (32%), Positives = 26/67 (37%)
Frame = +1
Query: 464 GGQGSPSGGGGGGGGRVHFHWSHIPVGDEYITLASVEGSIITGGGFGGGQGLPGKNGSIS 523
GG+G GGG GGGG H G G GGG+G G G S
Sbjct: 304 GGEGGGGGGGDGGGGGGDGH----------------------GSGGGGGEG-DGGGGGGS 414
Query: 524 GKACPKG 530
+ P+G
Sbjct: 415 PQVSPQG 435
>TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.33), partial
(12%)
Length = 565
Score = 34.3 bits (77), Expect = 0.12
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 19/92 (20%)
Frame = -2
Query: 438 GGGS--GGTVLLFVQTLALGDSSIISTVGGQGSPSGGGGGGG--------------GRVH 481
GGG+ GG V LF L + ++G + G GGGGG G
Sbjct: 564 GGGAVVGGVVELFEAEEILPAAGAAGSLG*EEGAVGDGGGGGGGGGENGCCCGGG*GGFE 385
Query: 482 FHWSHIPVGDEYITLASVEGSI---ITGGGFG 510
W +PVG+E + +G + + GGFG
Sbjct: 384 VEWDELPVGEELVGGEHGDGVLEAELEVGGFG 289
Score = 28.9 bits (63), Expect = 4.9
Identities = 25/74 (33%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Frame = -2
Query: 471 GGGGGGGGRVHFHWSH--IPVGDEYITLASVEGSIITGGGFGGGQGLPGKNGSISGKACP 528
GGG GG V + +P +L EG++ GGG GGG G+NG G
Sbjct: 564 GGGAVVGGVVELFEAEEILPAAGAAGSLG*EEGAVGDGGGGGGG---GGENGCCCGGG-- 400
Query: 529 KGLYGIFCEECPVG 542
G + + +E PVG
Sbjct: 399 *GGFEVEWDELPVG 358
>AV410198
Length = 337
Score = 34.3 bits (77), Expect = 0.12
Identities = 25/61 (40%), Positives = 28/61 (44%)
Frame = -2
Query: 464 GGQGSPSGGGGGGGGRVHFHWSHIPVGDEYITLASVEGSIITGGGFGGGQGLPGKNGSIS 523
GG G SGGGG GGG V VGD S++G + G G G G P N I
Sbjct: 279 GGGGIGSGGGGWGGGLVGEAEGAEFVGD------SLDGGVWESSGLGVGGGAP--NDEIG 124
Query: 524 G 524
G
Sbjct: 123 G 121
Score = 30.8 bits (68), Expect = 1.3
Identities = 26/85 (30%), Positives = 36/85 (41%)
Frame = -2
Query: 310 IRSVKVEYSGVISASGLGCTGGLGKGRYFENGIGGGGGHGGYGGDGYYNGNFIEGGTTYG 369
+R V E+ S G GG+G G G G GGG G + G+ ++GG
Sbjct: 321 LRDVVGEWDEDFSGGG----GGIGSG-----GGGWGGGLVGEAEGAEFVGDSLDGGVWES 169
Query: 370 DVDLPCELGSGSGNDSIAGATAGGG 394
+G G+ ND I G+ G G
Sbjct: 168 S---GLGVGGGAPNDEIGGSGFGFG 103
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 33.9 bits (76), Expect = 0.15
Identities = 28/73 (38%), Positives = 33/73 (44%), Gaps = 17/73 (23%)
Frame = +2
Query: 324 SGLGCTGGLGKGRYFENGIGGGGGH---GGYGGDGYYNGNFIEGG--------------T 366
SG G +GG G+G F++G GGG G GG GGD G GG T
Sbjct: 152 SGGGRSGG-GRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGSGGGGRGGRGTPLRAAGKKT 328
Query: 367 TYGDVDLPCELGS 379
T+ D D C L S
Sbjct: 329 TFADED*GCWLAS 367
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,472,173
Number of Sequences: 28460
Number of extensions: 368358
Number of successful extensions: 5089
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 2920
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4292
length of query: 1162
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1062
effective length of database: 2,051,600
effective search space: 2178799200
effective search space used: 2178799200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144766.10


