

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.21 - phase: 0
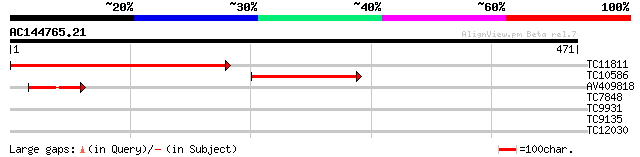
(471 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11811 similar to PIR|D96538|D96538 cytosolic tRNA-Ala syntheta... 342 8e-95
TC10586 similar to GB|CAA80380.1|1673365|ATTRNASYA mitochondrial... 159 9e-40
AV409818 48 3e-06
TC7848 homologue to UP|Q9AVR8 (Q9AVR8) Sucrose synthase isoform ... 28 3.2
TC9931 weakly similar to GB|AAO39953.1|28372942|BT003725 At5g152... 28 3.2
TC9135 homologue to UP|Q9SBJ1 (Q9SBJ1) Pyruvate dehydrogenase ki... 27 7.2
TC12030 27 7.2
>TC11811 similar to PIR|D96538|D96538 cytosolic tRNA-Ala synthetase
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (18%)
Length = 680
Score = 342 bits (877), Expect = 8e-95
Identities = 156/184 (84%), Positives = 166/184 (89%), Gaps = 1/184 (0%)
Frame = +2
Query: 1 MPETESIDIEWPAKRVRDTFAKFFEE-KNHVHWKSSAVVPLNDPTLLFANAGMNQFKPIF 59
MP +++D+EWPAKRVRDTF FF+E KNHV+W+SS VVP NDPTLLFANAGMNQFKPIF
Sbjct: 128 MPGPDALDLEWPAKRVRDTFVNFFKESKNHVYWQSSPVVPFNDPTLLFANAGMNQFKPIF 307
Query: 60 LGTVDPNTALSKLTRACNIQKCIRAGGKHNDLDDVGKDTYHHTFFEMLGNWSFGDYFKEE 119
LGT DPNT LSKL+RACN QKCIRAGGKHNDLDDVGKDTYHHTFFEMLGNWSFGDYFK E
Sbjct: 308 LGTADPNTDLSKLSRACNTQKCIRAGGKHNDLDDVGKDTYHHTFFEMLGNWSFGDYFKTE 487
Query: 120 AISWAWELLTEVYNLPSDRIYVTYFGGDEKTGLGPDNEARDIWLKFLPPGRVLPFGSKDN 179
AI WAWELLT+VY LPSDRIY TYFGGDEK GL PD EARDIWL+FLPPGRVLPFG KDN
Sbjct: 488 AIGWAWELLTKVYKLPSDRIYATYFGGDEKAGLAPDTEARDIWLQFLPPGRVLPFGCKDN 667
Query: 180 FWEM 183
FWEM
Sbjct: 668 FWEM 679
>TC10586 similar to GB|CAA80380.1|1673365|ATTRNASYA mitochondrial tRNA-Ala
synthetase {Arabidopsis thaliana;} , partial (9%)
Length = 644
Score = 159 bits (402), Expect = 9e-40
Identities = 76/91 (83%), Positives = 85/91 (92%)
Frame = +1
Query: 202 NRNASSFVNDEDPNCIELWNLVFIQFNREADGSLKSLPAKHVDTGMGFERLTSILQHKNS 261
NR+A+S VN++DP CIE+WNLVFIQFNREADGSLKSLPAKHVDTGMGFERLTSILQ++ S
Sbjct: 1 NRDAASLVNNDDPTCIEIWNLVFIQFNREADGSLKSLPAKHVDTGMGFERLTSILQNQMS 180
Query: 262 NYDTDVFVPIFDAIQQATGARPYSGKVGPHD 292
+YDTDVF+PIFDAIQ ATGARPYSGKVG D
Sbjct: 181 HYDTDVFLPIFDAIQLATGARPYSGKVGA*D 273
>AV409818
Length = 426
Score = 48.1 bits (113), Expect = 3e-06
Identities = 22/48 (45%), Positives = 33/48 (67%)
Frame = +2
Query: 16 VRDTFAKFFEEKNHVHWKSSAVVPLNDPTLLFANAGMNQFKPIFLGTV 63
+RD F +F+ + H S++++P +DPT+L AGM QFKPIFLG +
Sbjct: 266 IRDRFLRFYASRGHKVLPSASLIP-DDPTVLLTIAGMLQFKPIFLGKI 406
>TC7848 homologue to UP|Q9AVR8 (Q9AVR8) Sucrose synthase isoform 3 ,
complete
Length = 2779
Score = 28.1 bits (61), Expect = 3.2
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 1/27 (3%)
Frame = -1
Query: 423 GFPLDLTQLMAEEKKF-SVDIEGFHRA 448
GFP Q + EE KF S D +GFH+A
Sbjct: 199 GFPCPWLQSLTEESKFHSFDQQGFHQA 119
>TC9931 weakly similar to GB|AAO39953.1|28372942|BT003725 At5g15290
{Arabidopsis thaliana;}, partial (60%)
Length = 609
Score = 28.1 bits (61), Expect = 3.2
Identities = 12/31 (38%), Positives = 19/31 (60%)
Frame = -1
Query: 395 IRKFNKAVEHVQGNILSGEATFELSHTFGFP 425
++K N ++EH NI S +A + SHT +P
Sbjct: 510 VKKTNTSIEHQTENI*STQAKWNTSHTHFYP 418
>TC9135 homologue to UP|Q9SBJ1 (Q9SBJ1) Pyruvate dehydrogenase kinase ,
partial (31%)
Length = 538
Score = 26.9 bits (58), Expect = 7.2
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = +1
Query: 132 YNLPSDRIYVTYFGGD 147
Y +P R+Y YFGGD
Sbjct: 220 YGIPISRLYARYFGGD 267
>TC12030
Length = 401
Score = 26.9 bits (58), Expect = 7.2
Identities = 16/57 (28%), Positives = 25/57 (43%)
Frame = -3
Query: 77 NIQKCIRAGGKHNDLDDVGKDTYHHTFFEMLGNWSFGDYFKEEAISWAWELLTEVYN 133
NIQ+ + G + DD+ D + + NW G F E+ +S L E +N
Sbjct: 357 NIQQ*SKRGNHNTMADDITSDIMNK-IVPLTSNWITGSSFNEQPVSTEHILKIEQFN 190
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,178,256
Number of Sequences: 28460
Number of extensions: 113907
Number of successful extensions: 431
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 430
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 430
length of query: 471
length of database: 4,897,600
effective HSP length: 94
effective length of query: 377
effective length of database: 2,222,360
effective search space: 837829720
effective search space used: 837829720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144765.21


