

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.7 + phase: 0
(228 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
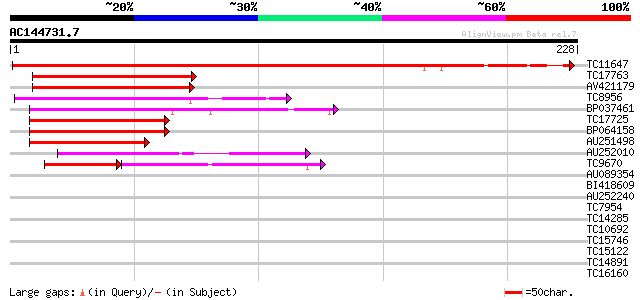
Score E
Sequences producing significant alignments: (bits) Value
TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, pa... 221 1e-58
TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor,... 76 4e-15
AV421179 75 1e-14
TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, pa... 75 1e-14
BP037461 71 2e-13
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 70 4e-13
BP064158 67 2e-12
AU251498 63 4e-11
AU252010 56 5e-09
TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%) 38 1e-08
AU089354 38 0.002
BI418609 27 2.9
AU252240 27 2.9
TC7954 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein... 27 3.8
TC14285 similar to UP|Q9AXU5 (Q9AXU5) Pathogen-inducible alpha-d... 26 5.0
TC10692 weakly similar to UP|BAD08916 (BAD08916) Calcium-binding... 26 5.0
TC15746 26 6.5
TC15122 similar to UP|Q94AY5 (Q94AY5) At1g19950/T20H2_25, partia... 26 6.5
TC14891 25 8.5
TC16160 25 8.5
>TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, partial (25%)
Length = 843
Score = 221 bits (562), Expect = 1e-58
Identities = 121/242 (50%), Positives = 156/242 (64%), Gaps = 16/242 (6%)
Frame = +1
Query: 2 SSVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKV 61
+S RK GRQKIEMKK++NESNLQVTFSK +GLFKKASELC LC ++ALVVFSP KV
Sbjct: 67 TSRRKSLGRQKIEMKKMTNESNLQVTFSKRRSGLFKKASELCILCDVEIALVVFSPGEKV 246
Query: 62 FSFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGD 121
FSFGHP+++ VI R+ S P Q TMQ+IEA RN+ V ELN +L+QINN LD + G+
Sbjct: 247 FSFGHPSVEAVIKRYFSQAPPQTSDTMQYIEAQRNSKVHELNVELSQINNLLDTVRNHGE 426
Query: 122 ELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQH-------------AAT 168
EL+ LHK +A+FWWAC ++ M+R +LE F+ AL ELKK + ++ AT
Sbjct: 427 ELNRLHKAAQAQFWWACQIEEMDRPKLEQFRMALNELKKQISRYCDRTRIHGAPAASTAT 606
Query: 169 RTLP---FFVGNTSSSNIYLHHQPNPQQAEMFPPQIFQNAMLQLQTHFFDGSMIPHHVFN 225
P FF +S+ I HH +P ++F PQ F+ +M Q H F G +FN
Sbjct: 607 TNPPTHMFFASGSSNVMIPPHHPQSP-PPQVFAPQFFEGSMKQ-GHHLFGG------IFN 762
Query: 226 NM 227
NM
Sbjct: 763 NM 768
>TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor, partial
(35%)
Length = 737
Score = 76.3 bits (186), Expect = 4e-15
Identities = 36/66 (54%), Positives = 49/66 (73%)
Frame = +3
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R+KI++KKI N + QVTFSK GLFKKA EL LC ADVALVVFS +GK+F + + ++
Sbjct: 507 REKIQIKKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALVVFSSTGKLFEYSNLSM 686
Query: 70 DTVIDR 75
+++R
Sbjct: 687 KEILER 704
>AV421179
Length = 207
Score = 74.7 bits (182), Expect = 1e-14
Identities = 35/65 (53%), Positives = 48/65 (73%)
Frame = +3
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R++I++KKI N S+ QVTFSK GLFKKA EL TLC AD+AL+VFS + K+F + ++
Sbjct: 12 RKRIQIKKIDNISSRQVTFSKRRKGLFKKAQELSTLCDADIALIVFSATNKLFEYASSSI 191
Query: 70 DTVID 74
VI+
Sbjct: 192QKVIE 206
>TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, partial
(58%)
Length = 1100
Score = 74.7 bits (182), Expect = 1e-14
Identities = 45/113 (39%), Positives = 66/113 (57%), Gaps = 2/113 (1%)
Frame = +2
Query: 3 SVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVF 62
S+R+ GR KIE+K I N +N QVT+SK NG+FKKA EL LC A V+L++FS + K+
Sbjct: 122 SLREIMGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMH 301
Query: 63 SFGHPNLDT--VIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTL 113
+ P L T +ID++ + G + +H + L +L +INN L
Sbjct: 302 EYISPGLTTKRIIDQYQKTL-----GDIDLWRSHYEKMLENLK-KLKEINNKL 442
>BP037461
Length = 551
Score = 70.9 bits (172), Expect = 2e-13
Identities = 44/129 (34%), Positives = 71/129 (54%), Gaps = 5/129 (3%)
Frame = +2
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF-GHP 67
GR ++E+K+I N+ N QVTF+K NGL KKA EL LC A+VAL++FS GK++ F
Sbjct: 11 GRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSSS 190
Query: 68 NLDTVIDRFLSL---IPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELS 124
++ ++R+ P N T + +E +L A+ + + +G++L
Sbjct: 191 SMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRS--QRNLMGEDLG 364
Query: 125 NLH-KETEA 132
L+ KE E+
Sbjct: 365 PLNSKELES 391
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 69.7 bits (169), Expect = 4e-13
Identities = 32/56 (57%), Positives = 42/56 (74%)
Frame = +3
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
GR ++E+K+I N+ N QVTF+K NGL KKA EL LC A+VAL+VFS GK++ F
Sbjct: 279 GRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSTRGKLYEF 446
>BP064158
Length = 394
Score = 67.4 bits (163), Expect = 2e-12
Identities = 32/56 (57%), Positives = 40/56 (71%)
Frame = +3
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
GR K++ +K+ N +N QVTFSK NGL KKA EL LC DVAL++FSPSG+ F
Sbjct: 168 GRVKLQNQKVENTTNRQVTFSKRRNGLIKKAYELSVLCXFDVALIMFSPSGRACLF 335
>AU251498
Length = 342
Score = 63.2 bits (152), Expect = 4e-11
Identities = 31/48 (64%), Positives = 36/48 (74%)
Frame = +3
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFS 56
GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS
Sbjct: 48 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS 191
>AU252010
Length = 328
Score = 56.2 bits (134), Expect = 5e-09
Identities = 33/102 (32%), Positives = 52/102 (50%)
Frame = +1
Query: 20 NESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNLDTVIDRFLSL 79
N SN QVT+SK NG+ KKA E+ LC A V+L++F+ SGK+ + P+ T++D
Sbjct: 4 NSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISPS-TTLVD----- 165
Query: 80 IPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGD 121
H+ + R +A+ +N ++ KK D
Sbjct: 166 ---------MLERYHKTSGKRLWDAKHENLNGEIERLKKEND 264
>TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%)
Length = 1060
Score = 37.7 bits (86), Expect(2) = 1e-08
Identities = 18/31 (58%), Positives = 21/31 (67%)
Frame = +1
Query: 15 MKKISNESNLQVTFSKHHNGLFKKASELCTL 45
+K+I N +N QVTF K NGL KKA EL L
Sbjct: 1 IKRIENTTNRQVTFCKRRNGLLKKAYELSVL 93
Score = 36.6 bits (83), Expect(2) = 1e-08
Identities = 21/83 (25%), Positives = 45/83 (53%), Gaps = 1/83 (1%)
Frame = +3
Query: 46 CGADVALVVFSPSGKVFSFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQ 105
C A+VAL+VFS G+++ + + ++ IDR+ + + G EA+ +E +
Sbjct: 93 CDAEVALIVFSSRGRLYEYANNSVKATIDRYKKAC-SDSSGAGSASEANAQFYQQEADKL 269
Query: 106 LTQINNTLDAEKK-IGDELSNLH 127
QI+N + ++ +G+ L +++
Sbjct: 270 RVQISNLQNNNRQMMGESLGSMN 338
>AU089354
Length = 178
Score = 37.7 bits (86), Expect = 0.002
Identities = 16/39 (41%), Positives = 25/39 (64%)
Frame = +3
Query: 34 GLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNLDTV 72
G+ KKA E+ LC A V+L+ F+ SGK+ + P++ V
Sbjct: 3 GILKKAIEITVLCXAQVSLIXFAASGKMHDYIRPSITXV 119
>BI418609
Length = 538
Score = 26.9 bits (58), Expect = 2.9
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Frame = +2
Query: 186 HHQPNPQ--QAEMFPPQIFQNAMLQLQTHFFDGSMIPHHVFNNMV 228
HHQ Q Q + P +FQ + + H F S IPHH F++ V
Sbjct: 149 HHQQR*QCRQEALRPRDLFQASPCFRRHHRF--SQIPHHAFDSTV 277
>AU252240
Length = 398
Score = 26.9 bits (58), Expect = 2.9
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 2/30 (6%)
Frame = -3
Query: 186 HHQP--NPQQAEMFPPQIFQNAMLQLQTHF 213
HH+P +PQ E + P F +L L +HF
Sbjct: 375 HHEP*PSPQSFEYYRPMSFARPLLFLPSHF 286
>TC7954 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein, partial
(75%)
Length = 1708
Score = 26.6 bits (57), Expect = 3.8
Identities = 15/36 (41%), Positives = 21/36 (57%), Gaps = 4/36 (11%)
Frame = +2
Query: 158 LKKLVIQHAATRTLPF----FVGNTSSSNIYLHHQP 189
LKK I+ T+ +PF + N SSS+ +HHQP
Sbjct: 23 LKKKAIE---TKPVPFSSIRYKNNPSSSSFIIHHQP 121
>TC14285 similar to UP|Q9AXU5 (Q9AXU5) Pathogen-inducible
alpha-dioxygenase, partial (97%)
Length = 2266
Score = 26.2 bits (56), Expect = 5.0
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = -2
Query: 13 IEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGA 48
+ M+ SNES+ + +SKHH G F C+ C A
Sbjct: 93 VMMQNFSNESSNGICYSKHH-GCF------CSFCDA 7
>TC10692 weakly similar to UP|BAD08916 (BAD08916) Calcium-binding EF-hand
family protein-like, partial (32%)
Length = 634
Score = 26.2 bits (56), Expect = 5.0
Identities = 28/111 (25%), Positives = 47/111 (42%), Gaps = 16/111 (14%)
Frame = +2
Query: 73 IDRFLSLIPTQNDG------------TMQFIEAHRNANVRELNAQLTQIN----NTLDAE 116
+D + + NDG +M+ IEAH +V QLT+I +T D +
Sbjct: 101 VDEQFAALDLNNDGVLSRAELRTAFESMRLIEAHFGIDVATPPEQLTKIYDSVFDTFDGD 280
Query: 117 KKIGDELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAA 167
+ L +E K V DG+ + +++ ALE+ ++Q AA
Sbjct: 281 G--SGTVDRLEFRSEIKKIMLAVADGLGTNPIQM---ALEDDSNNLLQKAA 418
>TC15746
Length = 619
Score = 25.8 bits (55), Expect = 6.5
Identities = 21/81 (25%), Positives = 34/81 (41%)
Frame = +2
Query: 82 TQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLHKETEAKFWWACVVD 141
T+N G M+ N+ E + +L N +D++ + E EA
Sbjct: 38 TENVGDMKSENGDVQLNLAEDSEELKSKLNIMDSKLR----------EAEATIMMLNEER 187
Query: 142 GMNRDQLEIFKKALEELKKLV 162
MN + + FK+ LE LKK +
Sbjct: 188 RMNTKEKDFFKRELEVLKKKI 250
>TC15122 similar to UP|Q94AY5 (Q94AY5) At1g19950/T20H2_25, partial (56%)
Length = 1343
Score = 25.8 bits (55), Expect = 6.5
Identities = 11/40 (27%), Positives = 20/40 (49%)
Frame = -1
Query: 25 QVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
Q++F+ H N +K + T C + +P K+F+F
Sbjct: 371 QLSFTVHRNP*YKSITNSLTYCQNSHQNPILAPKAKLFNF 252
>TC14891
Length = 1172
Score = 25.4 bits (54), Expect = 8.5
Identities = 12/35 (34%), Positives = 18/35 (51%), Gaps = 4/35 (11%)
Frame = +1
Query: 17 KISNESNLQVT----FSKHHNGLFKKASELCTLCG 47
K+ + LQ T F H G+ + ++LCT CG
Sbjct: 190 KLIQDCKLQYTKSLVFQTHQLGILSQHTKLCTSCG 294
>TC16160
Length = 1164
Score = 25.4 bits (54), Expect = 8.5
Identities = 16/45 (35%), Positives = 20/45 (43%)
Frame = -3
Query: 178 TSSSNIYLHHQPNPQQAEMFPPQIFQNAMLQLQTHFFDGSMIPHH 222
TS S +HHQ NP+ A +QT F + S I HH
Sbjct: 223 TSLSPYPIHHQKNPRSHP-------APASYPVQTAFSETSQIDHH 110
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.133 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,970,379
Number of Sequences: 28460
Number of extensions: 54409
Number of successful extensions: 372
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 371
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 372
length of query: 228
length of database: 4,897,600
effective HSP length: 87
effective length of query: 141
effective length of database: 2,421,580
effective search space: 341442780
effective search space used: 341442780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC144731.7


