

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.5 + phase: 0 /pseudo
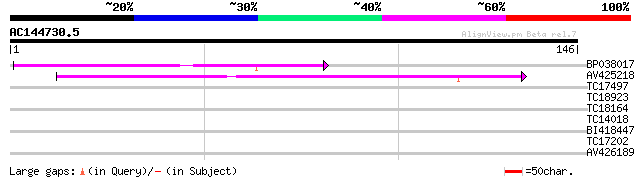
(146 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP038017 44 1e-05
AV425218 39 3e-04
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 34 0.012
TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1,... 33 0.026
TC18164 similar to GB|AAN33186.1|23452834|AF543710 elongation de... 25 4.1
TC14018 similar to UP|Q43465 (Q43465) Protein kinase 2, partial ... 25 4.1
BI418447 25 5.3
TC17202 similar to UP|Q9FQ10 (Q9FQ10) Sucrose-phosphatase , par... 25 7.0
AV426189 24 9.1
>BP038017
Length = 570
Score = 43.5 bits (101), Expect = 1e-05
Identities = 26/82 (31%), Positives = 39/82 (46%), Gaps = 1/82 (1%)
Frame = +2
Query: 2 DSTYKTNMYKMPMFEVVGVTLTDLTYSVGFGFETHEKEENFVWVLKMLRKLLTSKMNMPK 61
D+ Y+ N Y++P GV G E E +F W + R L++ + P
Sbjct: 329 DTMYRPNQYQVPFAPFTGVNHHGQNVLFGCALLLDESESSFTW---LFRTWLSAMNDRPP 499
Query: 62 V-IVTDRDMSLMKEVANVFPES 82
V I TD+D ++ VA VFPE+
Sbjct: 500 VSITTDQDRAIQAAVAQVFPET 565
>AV425218
Length = 419
Score = 39.3 bits (90), Expect = 3e-04
Identities = 28/122 (22%), Positives = 55/122 (44%), Gaps = 1/122 (0%)
Frame = +2
Query: 13 PMFEVVGVTLTDLTYSVGFGFETHEKEENFVWVLKMLRKLLTSKMNMPKVIVTDRDMSLM 72
P+ +VGV + G + E E+FVW+ + L ++ P+ I+TD ++
Sbjct: 5 PLATLVGVNHHGQSVLFGCALLSSEDSESFVWLFQSLLHCMSGV--PPQGIITDHSEAMK 178
Query: 73 KEVANVFPESYAMNCYFHVQANVKQRCVLDCKYPLGKKDGKE-VKPRDVVNKIMRAWKSM 131
K + V P + C ++ + Q+ + +Y + + V V+++ R WK +
Sbjct: 179 KAIETVLPSTRHRWCLSYIMKKLPQKLLGYAQYESIRHHLQNVVYDAVVIDEFERNWKKI 358
Query: 132 VE 133
VE
Sbjct: 359 VE 364
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 33.9 bits (76), Expect = 0.012
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 10/104 (9%)
Frame = +1
Query: 46 LKMLRKLLTSKMN-------MPKVIVTDRDMSLMKEVANVF---PESYAMNCYFHVQANV 95
LK LL +MN + ++ + D+ + K V + P YA++ F ++++V
Sbjct: 2032 LKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDV 2211
Query: 96 KQRCVLDCKYPLGKKDGKEVKPRDVVNKIMRAWKSMVESPTQEL 139
V+ + GKK G+ P +N + AW+ +E EL
Sbjct: 2212 FSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEERPLEL 2343
>TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1, complete
Length = 2061
Score = 32.7 bits (73), Expect = 0.026
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 10/104 (9%)
Frame = +1
Query: 46 LKMLRKLLTSKMN-------MPKVIVTDRDMSLMKEVANVF---PESYAMNCYFHVQANV 95
LK LL ++MN + ++ + D+ + K V + P YA++ F ++++V
Sbjct: 1495 LKTSNILLDNEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDV 1674
Query: 96 KQRCVLDCKYPLGKKDGKEVKPRDVVNKIMRAWKSMVESPTQEL 139
V+ + GKK K P +N + AW+ +E EL
Sbjct: 1675 FSFGVIVLEIISGKKIRKFYDPHHHLNLLSHAWRLWIEGSPLEL 1806
>TC18164 similar to GB|AAN33186.1|23452834|AF543710 elongation defective 1
{Arabidopsis thaliana;}, partial (31%)
Length = 1023
Score = 25.4 bits (54), Expect = 4.1
Identities = 22/92 (23%), Positives = 40/92 (42%), Gaps = 6/92 (6%)
Frame = +1
Query: 40 ENFVW-----VLKMLRKLLTSKMNMPKVIVTD-RDMSLMKEVANVFPESYAMNCYFHVQA 93
E+ VW LK+LRK + +++ P VIV R+ + V P + + +
Sbjct: 355 EHVVWGDKDLKLKLLRKGILTRIYAPMVIVQSLRESGVFSSVIASAPTLSKESFLQSIDS 534
Query: 94 NVKQRCVLDCKYPLGKKDGKEVKPRDVVNKIM 125
+ R V P +K G+ + + V K++
Sbjct: 535 SNSSRAVASVSLP-SRKAGRTXESQATVRKVL 627
>TC14018 similar to UP|Q43465 (Q43465) Protein kinase 2, partial (27%)
Length = 559
Score = 25.4 bits (54), Expect = 4.1
Identities = 21/63 (33%), Positives = 32/63 (50%)
Frame = +1
Query: 43 VWVLKMLRKLLTSKMNMPKVIVTDRDMSLMKEVANVFPESYAMNCYFHVQANVKQRCVLD 102
+W+LKM+RKL M V + D L +A V+ Y+M+ Y H+ + +RC D
Sbjct: 328 LWMLKMMRKL------MLVVTMNQCDRCLHW*MACVYKIIYSMH-YLHLSL-LFRRCSKD 483
Query: 103 CKY 105
Y
Sbjct: 484 IHY 492
>BI418447
Length = 443
Score = 25.0 bits (53), Expect = 5.3
Identities = 10/31 (32%), Positives = 19/31 (61%)
Frame = +2
Query: 43 VWVLKMLRKLLTSKMNMPKVIVTDRDMSLMK 73
+WV KML+K L +++ +P+ + T +K
Sbjct: 167 IWVSKMLQKSLLNRVQLPQSL*TQSHCGKIK 259
>TC17202 similar to UP|Q9FQ10 (Q9FQ10) Sucrose-phosphatase , partial (28%)
Length = 655
Score = 24.6 bits (52), Expect = 7.0
Identities = 13/48 (27%), Positives = 24/48 (49%)
Frame = +1
Query: 59 MPKVIVTDRDMSLMKEVANVFPESYAMNCYFHVQANVKQRCVLDCKYP 106
+P+ +VT +++M + N F + + HV+ + R L CK P
Sbjct: 385 VPQCLVTPFAINVM*SMTN*FQHLNFLKNFHHVKVSYIYRFELHCKSP 528
>AV426189
Length = 408
Score = 24.3 bits (51), Expect = 9.1
Identities = 10/55 (18%), Positives = 26/55 (47%)
Frame = +2
Query: 25 LTYSVGFGFETHEKEENFVWVLKMLRKLLTSKMNMPKVIVTDRDMSLMKEVANVF 79
+TY ++++ +W K+L + +++ KVI D D + ++ ++
Sbjct: 110 ITYKWPTWLHKQKEKQRIIWAYKILFLDVIFPLSLEKVIFVDADQVVRADMGELY 274
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,276,785
Number of Sequences: 28460
Number of extensions: 25352
Number of successful extensions: 126
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 125
length of query: 146
length of database: 4,897,600
effective HSP length: 82
effective length of query: 64
effective length of database: 2,563,880
effective search space: 164088320
effective search space used: 164088320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 51 (24.3 bits)
Medicago: description of AC144730.5


