

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.4 + phase: 0
(566 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
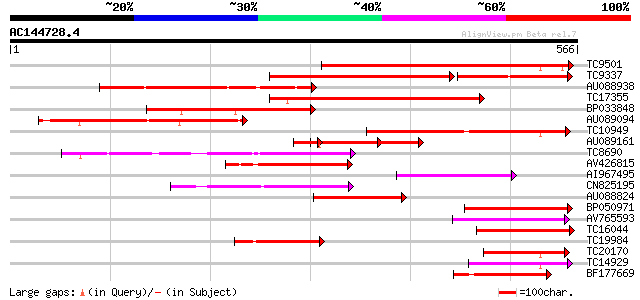
Score E
Sequences producing significant alignments: (bits) Value
TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesteras... 328 1e-90
TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B pre... 189 1e-73
AU088938 223 6e-59
TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%) 202 1e-52
BP033848 198 2e-51
AU089094 186 9e-48
TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precu... 184 3e-47
AU089161 96 8e-42
TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precu... 151 2e-37
AV426815 108 3e-24
AI967495 102 2e-22
CN825195 101 3e-22
AU088824 100 8e-22
BP050971 99 1e-21
AV765593 94 5e-20
TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 93 1e-19
TC19984 similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase , pa... 92 2e-19
TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isof... 92 2e-19
TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase ... 91 5e-19
BF177669 89 1e-18
>TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase ,
partial (29%)
Length = 934
Score = 328 bits (842), Expect = 1e-90
Identities = 158/259 (61%), Positives = 199/259 (76%), Gaps = 7/259 (2%)
Frame = +2
Query: 312 PTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSA 371
P KTIITG+KNF DGVKT+QTATF+ A GFIAKA+ FENTAG N HQAVALR QGD+SA
Sbjct: 5 PAKTIITGRKNFADGVKTMQTATFANQAPGFIAKAITFENTAGPNGHQAVALRNQGDQSA 184
Query: 372 FFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNII 431
F C I GYQDTLY +RQFYRNC ISGT+DFIFG + T+IQ+S IVVRKP NQ N +
Sbjct: 185 FVGCHILGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSPTLIQHSIIVVRKPNDNQFNTV 364
Query: 432 VADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQ 491
ADGT KNMPTG+V+Q+CEI+PE AL P R K++S+L RPWK YSR + ME+T+GD I
Sbjct: 365 TADGTPTKNMPTGIVIQDCEIVPEAALFPVRFKIKSYLGRPWKEYSRTVVMESTLGDFIH 544
Query: 492 PDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLSKADATKYTAAQWIEGG 549
P+G+ PWAG+ LDT ++AE+ANTGPG+N R+KW +GV+S+A+AT++TA ++++ G
Sbjct: 545 PEGWFPWAGSFALDTLYYAEHANTGPGANTAGRIKWKGYRGVISRAEATQFTAGEFLKAG 724
Query: 550 V-----WLPATGIPFDLGF 563
W+ A +P LGF
Sbjct: 725 PRSGLDWIKALHVPHYLGF 781
>TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (54%)
Length = 1145
Score = 189 bits (481), Expect(2) = 1e-73
Identities = 93/185 (50%), Positives = 122/185 (65%)
Frame = +2
Query: 260 DGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITG 319
DGSG + TV++A+ + P R+VIYVK GVY+EY +I K K NI++ GDG T+I+G
Sbjct: 29 DGSGNYTTVMEAVLAAPDYSMKRFVIYVKKGVYEEYAEIKKKKWNIMMVGDGMDVTVISG 208
Query: 320 KKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRG 379
++F DG T ++ATF+ GFIA+ + F+NTAG KHQAVALR D + FF C+I G
Sbjct: 209 NRSFGDGWTTFKSATFAVSGRGFIARGITFQNTAGPEKHQAVALRSDSDLAVFFRCSITG 388
Query: 380 YQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQK 439
YQD+LY H RQFY+ C I+GTVDFIFG A+ V QN I RK +Q+N I A G
Sbjct: 389 YQDSLYPHTMRQFYKECRITGTVDFIFGDATAVFQNCNIQARKGLQDQKNTITAQGRKDP 568
Query: 440 NMPTG 444
+G
Sbjct: 569 GQSSG 583
Score = 104 bits (259), Expect(2) = 1e-73
Identities = 50/116 (43%), Positives = 73/116 (62%), Gaps = 1/116 (0%)
Frame = +3
Query: 448 QNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTC 507
Q C I + L P ++++L RPWK YSR +FM++ + D++ G+L W T F DT
Sbjct: 594 QYCNITADSELLPSVGSIQTYLGRPWKEYSRTVFMQSFMSDVVSSQGWLEW-NTSFDDTL 770
Query: 508 FFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
F+ EY N G G+ V RVKW G VL+ ++A K+T Q+IEG +WLP+TG+ + G
Sbjct: 771 FYGEYLNYGAGAGVGNRVKWGGYHVLNDSEAGKFTVNQFIEGNLWLPSTGVAYTGG 938
>AU088938
Length = 641
Score = 223 bits (568), Expect = 6e-59
Identities = 121/217 (55%), Positives = 154/217 (70%)
Frame = +1
Query: 90 KAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAAD 149
+AFNM+D+L VE+E +N KMAL+DCKDLL+ AI ELQAS +L + +VN R+A+
Sbjct: 7 EAFNMSDRLTVEHENSNPGVKMALEDCKDLLQSAIQELQASGVLV-QXXXLLDVNQRSAE 183
Query: 150 LKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAAL 209
LKNWL AV AYQQSCLDGFDT GEK VQ +LQ GSLD VGKLT LALD+V+ I+KVL A
Sbjct: 184 LKNWLSAVVAYQQSCLDGFDTKGEKXVQDKLQQGSLDDVGKLTGLALDIVSGISKVLGAF 363
Query: 210 DLDLNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVL 269
D +L++KP +RRL E+D +G P W+S ADRKL+ S +V PNA+VAKDG G FK +
Sbjct: 364 DFNLHLKP-ARRLLEIDAEGYPTWLSFADRKLVESKS---NVLPNAIVAKDGXGXFKNIT 531
Query: 270 DAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNIL 306
+AINSY + +AGVYD+Y + K + L
Sbjct: 532 EAINSYLXXTXVNPHL-CQAGVYDKYSTLTKXTPHSL 639
>TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%)
Length = 848
Score = 202 bits (514), Expect = 1e-52
Identities = 102/218 (46%), Positives = 134/218 (60%), Gaps = 3/218 (1%)
Frame = +1
Query: 260 DGSGKFKTVLDAINSYP---KNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTI 316
+G+ + ++ DAI + P K G ++IYV+ G Y+EY+ + K KKNIL+ GDG KTI
Sbjct: 172 NGTDNYTSIGDAIAAAPNHTKPQDGYFLIYVREGYYEEYVIVPKEKKNILLVGDGINKTI 351
Query: 317 ITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCA 376
ITG + +DG T ++TF+ E F A + F NTAG KHQAVA+R D S F+ C+
Sbjct: 352 ITGNHSVIDGWTTFNSSTFAVSGERFTAVDVTFRNTAGPAKHQAVAVRNNADLSTFYRCS 531
Query: 377 IRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGT 436
GYQDTLY H+ RQFYR C+I GTVDFIFG A+ V QN I RKP Q+N + A G
Sbjct: 532 FEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAAVVFQNCNIYARKPLPFQKNAVTAQGR 711
Query: 437 VQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWK 474
N TG+ +QNC I P L + S+L RPW+
Sbjct: 712 TDPNQNTGISIQNCTIDAAPDLAAELNSTLSYLGRPWE 825
>BP033848
Length = 525
Score = 198 bits (503), Expect = 2e-51
Identities = 96/174 (55%), Positives = 128/174 (73%), Gaps = 5/174 (2%)
Frame = +2
Query: 137 NSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDT--DGEKQVQSQLQTGSLDHVGKLTAL 194
++++ V+++ D +NWL AV +YQQSC++GF+ GE+ V+ QLQ GSLD +GKLT +
Sbjct: 2 DNNIQAVHEQTPDFRNWLSAVMSYQQSCMEGFEDGKSGEEAVKGQLQEGSLDQMGKLTGI 181
Query: 195 ALDVVTAITKVLAALDLDLNVKPSSRRLFE---VDEDGNPEWMSGADRKLLADMSTGMSV 251
ALD+V+ ++++L DL L++ P+SRRL E VD +G P W SGADRKLL M G V
Sbjct: 182 ALDIVSDLSRILQTFDLKLDLNPASRRLLEAEEVDHEGLPSWFSGADRKLLGKMKKGGKV 361
Query: 252 TPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNI 305
TPNAVVAKDGSGKFKT+ AI SYPK +GRY+IYV+AGVYDEYI + K N+
Sbjct: 362 TPNAVVAKDGSGKFKTIASAIASYPKGFKGRYIIYVRAGVYDEYITVPKQAVNL 523
>AU089094
Length = 639
Score = 186 bits (472), Expect = 9e-48
Identities = 97/214 (45%), Positives = 140/214 (65%), Gaps = 5/214 (2%)
Frame = +1
Query: 29 NNGPADANNGGELTSHTKAVTAVCQNSDDHKFCADTLGSV---NTSDPNDYIKAVVKTSI 85
N+ PA E+ + ++V +CQN+DD K C DTL SV + +DP YI VK +
Sbjct: 1 NSAPA------EVQTQQRSVRTICQNTDDQKLCHDTLSSVKGVDAADPKAYIATAVKAAS 162
Query: 86 ESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSILAADNSSVHNVND 145
+SVIKAFNM+D+L E + KMALDDCKDLL+ A+ L S+ L DN ++ V++
Sbjct: 163 DSVIKAFNMSDRLTTEYGNKDHGIKMALDDCKDLLQSAMQSLDLSTTLVRDN-NIQGVHN 339
Query: 146 RAADLKNWLGAVFAYQQSCLDGF--DTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAIT 203
+ D +NWLGAV +YQQ+C++GF D DGEK ++ QL T S+DHV ++ A+ LD+VT +T
Sbjct: 340 QIPDFRNWLGAVISYQQACMEGFDNDXDGEKXIKEQLXTESMDHVXRIXAITLDIVTNMT 519
Query: 204 KVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGA 237
K+L +L L++KP+SRRL D D N ++ G+
Sbjct: 520 KILXQFNLKLDLKPASRRLLXXDVD-NEGYLHGS 618
>TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (34%)
Length = 773
Score = 184 bits (467), Expect = 3e-47
Identities = 93/207 (44%), Positives = 129/207 (61%), Gaps = 3/207 (1%)
Frame = +3
Query: 357 KHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNS 416
KHQAVA+R D+S F+ C+ G+QDTLYAH++RQFYR C+I+GT+DFIFG A+++ QN
Sbjct: 6 KHQAVAMRSGSDRSVFYRCSFDGFQDTLYAHSNRQFYRECDITGTIDFIFGNAASIFQNC 185
Query: 417 KIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAY 476
KI+ R+P NQ I A G N TG+V+Q I P ++L RPWK +
Sbjct: 186 KIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITP----LESTYTAPTYLGRPWKDF 353
Query: 477 SRAIFMENTIGDLIQPDGFLPW-AGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLS 533
S + M++ IG ++P G++ W A T F+AE+ N+GPGS+V RVKW K L+
Sbjct: 354 STTLIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLT 533
Query: 534 KADATKYTAAQWIEGGVWLPATGIPFD 560
D K+T A +I+G WLP T + FD
Sbjct: 534 ADDLGKFTLASFIQGPEWLPDTAVAFD 614
>AU089161
Length = 400
Score = 96.3 bits (238), Expect(2) = 8e-42
Identities = 48/74 (64%), Positives = 59/74 (78%), Gaps = 2/74 (2%)
Frame = +1
Query: 301 TKKNILI--YGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKH 358
T+KN++ G GP KT+ITG KNFVDG KT+++ATF+TVA F+AK+M FENTAGA+KH
Sbjct: 55 TRKNLIF*CTGXGPKKTVITGNKNFVDGYKTMRSATFTTVAPDFMAKSMGFENTAGASKH 234
Query: 359 QAVALRVQGDKSAF 372
QAVALRVQG F
Sbjct: 235 QAVALRVQGRPLGF 276
Score = 91.7 bits (226), Expect(2) = 8e-42
Identities = 40/46 (86%), Positives = 44/46 (94%)
Frame = +2
Query: 368 DKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVI 413
D+SAFFDCAI GYQDTLYAHAHRQFYRNCEISGTVDFIFGY++ +I
Sbjct: 263 DRSAFFDCAISGYQDTLYAHAHRQFYRNCEISGTVDFIFGYSTLLI 400
Score = 51.6 bits (122), Expect = 3e-07
Identities = 24/29 (82%), Positives = 25/29 (85%)
Frame = +3
Query: 284 VIYVKAGVYDEYIQIDKTKKNILIYGDGP 312
VIYVKAGVYDEYIQIDK K NIL+YG P
Sbjct: 9 VIYVKAGVYDEYIQIDKKKPNILMYGXWP 95
>TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor
(Pectinesterase) , partial (59%)
Length = 1082
Score = 151 bits (382), Expect = 2e-37
Identities = 103/302 (34%), Positives = 156/302 (51%), Gaps = 8/302 (2%)
Frame = +3
Query: 52 CQNSDDHKFCADTLGSVN------TSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKN 105
C+++ D C + V+ T DP I + T S I+ + K A++N N
Sbjct: 243 CEHALDASSCLAHVSEVSQSPISATKDPKLNILISLMTKSTSHIQEAMVKTK-AIKNRIN 419
Query: 106 NQSTKMALDDCKDLLEFAIDELQASSI-LAADNSSVHNVNDRAADLKNWLGAVFAYQQSC 164
N + AL DC+ L++ +ID + S + L DN+ H D WL V +C
Sbjct: 420 NPREEAALSDCEQLMDLSIDRVWDSVMALTKDNTDSHQ------DAHAWLSGVLTNHATC 581
Query: 165 LDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFE 224
LDG + ++++++ D+++ LA L L K + ++ +
Sbjct: 582 LDGLEGPSRALMEAEIE---------------DLISRSKTSLALLVSVLAPKGGNEQIID 716
Query: 225 VDEDGN-PEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRY 283
DG+ P W++ DR+LL + S G V N VVAKDGSG+FKTV +A+ S P + + RY
Sbjct: 717 EPLDGDFPSWVTRKDRRLL-ESSVG-DVNANVVVAKDGSGRFKTVAEAVASAPDSGKTRY 890
Query: 284 VIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFI 343
VIYVK G Y E I+I K K N+++ GDG TIITG N +DG T ++AT + V +GFI
Sbjct: 891 VIYVKKGTYKENIEIGKKKTNVMLTGDGMDATIITGNLNVIDGSTTFKSATVAAVGDGFI 1070
Query: 344 AK 345
A+
Sbjct: 1071AQ 1076
>AV426815
Length = 391
Score = 108 bits (269), Expect = 3e-24
Identities = 61/127 (48%), Positives = 78/127 (61%)
Frame = +3
Query: 216 KPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSY 275
K SR+L E E PEW+S ADR+LL +V + VVA DGSG FKTV A+ +
Sbjct: 24 KTGSRKLMEAGEVW-PEWISAADRRLLQ----AGTVKADVVVAADGSGNFKTVSAAVAAA 188
Query: 276 PKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATF 335
P+ + RYVI +KAGVY E +++ K K NI+ GDG T TIITG +N VDG T +AT
Sbjct: 189 PEKSKTRYVIKIKAGVYRENVEVPKKKTNIMFLGDGRTTTIITGSRNVVDGSTTFHSATV 368
Query: 336 STVAEGF 342
+ V F
Sbjct: 369 AVVGGNF 389
>AI967495
Length = 390
Score = 102 bits (253), Expect = 2e-22
Identities = 53/121 (43%), Positives = 69/121 (56%), Gaps = 1/121 (0%)
Frame = +1
Query: 387 HAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVV 446
HA RQFY+ C+I GTVD IFG A+ V QN KI +KP Q N+I A G TG+
Sbjct: 19 HAQRQFYKQCQIYGTVDLIFGNAAVVFQNCKIFAKKPLDGQANMITAQGRDDPFQNTGIT 198
Query: 447 LQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQF-LD 505
+ N EI P L+P K + L RPW+ YSR + ++ + L+ P G+ W T F LD
Sbjct: 199 IHNSEIRAAPDLKPVVNKHNTSLGRPWQQYSRVVVLKTFMDTLVSPLGWSSWDDTDFALD 378
Query: 506 T 506
T
Sbjct: 379 T 381
>CN825195
Length = 551
Score = 101 bits (252), Expect = 3e-22
Identities = 65/187 (34%), Positives = 103/187 (54%), Gaps = 4/187 (2%)
Frame = +2
Query: 161 QQSCLDGF-DTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKP-S 218
Q +C DGF DT G+ + Q + L ++V+ + A D + P
Sbjct: 26 QDTCADGFADTSGDVKDQMAVNLKDLS----------ELVSNCLAIFAGAGDDFSGVPIQ 175
Query: 219 SRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKN 278
+RRL + +D P W++ DR+LL+ + M + VV+KDG+G KT+ +AI P+
Sbjct: 176 NRRLLAMRDDNFPRWLNRRDRRLLSLPVSAMQA--DVVVSKDGNGTAKTIAEAIKKVPEY 349
Query: 279 HQGRYVIYVKAGVYDE-YIQIDKTKKNILIYGDGPTKTIITGKKNFVDG-VKTIQTATFS 336
R++IYV+AG Y+E ++I + + N++I GDG KT+ITG KN + G + T TA+F+
Sbjct: 350 GSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMYGLLTTFHTASFA 529
Query: 337 TVAEGFI 343
GFI
Sbjct: 530 ASGAGFI 550
>AU088824
Length = 421
Score = 100 bits (248), Expect = 8e-22
Identities = 53/93 (56%), Positives = 61/93 (64%)
Frame = +1
Query: 304 NILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVAL 363
NIL+YGDGPTKTIITG+KNF DGVKT+QTATF+ A GFIAKA+ FENTAG +
Sbjct: 10 NILMYGDGPTKTIITGRKNFADGVKTMQTATFANQAPGFIAKAITFENTAGPTVTKLWH* 189
Query: 364 RVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNC 396
+G +SAF A G L QFYR C
Sbjct: 190 XTKGXQSAFRWAAHLGLPKXLLVQTTXQFYRXC 288
>BP050971
Length = 477
Score = 99.4 bits (246), Expect = 1e-21
Identities = 47/110 (42%), Positives = 68/110 (61%), Gaps = 2/110 (1%)
Frame = -1
Query: 455 EPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPW-AGTQFLDTCFFAEYA 513
+P QP + +FL RPWK +SR +FME+ + ++I P+G+L W T + D FF EY
Sbjct: 477 DPDFQPSNGSIPTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETYTTYDDPLFFGEYP 298
Query: 514 NTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
N GPGS V RVKW G VL+ + A +T +I+G +WLP+ G+ + G
Sbjct: 297 NKGPGSGVANRVKWRGYHVLNNSPAMDFTVNPFIKGDLWLPSPGVSYHAG 148
>AV765593
Length = 501
Score = 94.4 bits (233), Expect = 5e-20
Identities = 43/118 (36%), Positives = 66/118 (55%), Gaps = 1/118 (0%)
Frame = -1
Query: 443 TGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQ 502
TG + NC I L ++L RPWK YSR ++M+ ++ ++I G+ W G
Sbjct: 495 TGTSIHNCTIRAADDLASSNGATATYLGRPWKEYSRTVYMQTSMDNVIDSAGWRAWDGEF 316
Query: 503 FLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPF 559
L T ++AE+ N+GPGSN RV W G V++ DA +T + ++ G WLP TG+ +
Sbjct: 315 ALSTLYYAEFNNSGPGSNTNGRVTWQGYHVINATDAANFTVSNFLLGDDWLPQTGVSY 142
>TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (43%)
Length = 564
Score = 92.8 bits (229), Expect = 1e-19
Identities = 45/100 (45%), Positives = 65/100 (65%), Gaps = 2/100 (2%)
Frame = +3
Query: 467 SFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVK 526
++L RPWK YSR ++M + IGD + G+L W T LDT ++ EY N GPG+ V RV
Sbjct: 30 TYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVN 209
Query: 527 W-GKGVLSKA-DATKYTAAQWIEGGVWLPATGIPFDLGFT 564
W G V++ A +A+++T AQ+I G WLP+TG+ F G +
Sbjct: 210 WPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGLS 329
>TC19984 similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase , partial (11%)
Length = 300
Score = 92.4 bits (228), Expect = 2e-19
Identities = 43/90 (47%), Positives = 58/90 (63%)
Frame = +3
Query: 225 VDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYV 284
+D DG P W+ DR+LL + PN VAKDGSG FKT+ +A+N P ++GRYV
Sbjct: 36 LDSDGFPSWVHVDDRRLLK--AADNRPKPNVTVAKDGSGDFKTISEALNKIPATYKGRYV 209
Query: 285 IYVKAGVYDEYIQIDKTKKNILIYGDGPTK 314
+YVK GVYDE + + K +N+ +YGDG K
Sbjct: 210 VYVKEGVYDEQVTVTKKMQNVTMYGDGSQK 299
>TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (32%)
Length = 592
Score = 92.4 bits (228), Expect = 2e-19
Identities = 43/88 (48%), Positives = 60/88 (67%), Gaps = 2/88 (2%)
Frame = +1
Query: 474 KAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGV 531
K YSR +F+++ + DLI P G+L W T LDT ++ EY N GPGSN ARVKWG + +
Sbjct: 1 KLYSRTVFLKSYMEDLIDPAGWLEWNDTFALDTLYYGEYQNRGPGSNPSARVKWGGYRVI 180
Query: 532 LSKADATKYTAAQWIEGGVWLPATGIPF 559
S +A+++T Q+I+G WL +TGIPF
Sbjct: 181 NSSTEASQFTVGQFIQGSDWLNSTGIPF 264
>TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase (Fragment)
, partial (24%)
Length = 693
Score = 90.9 bits (224), Expect = 5e-19
Identities = 42/106 (39%), Positives = 62/106 (57%), Gaps = 2/106 (1%)
Frame = +2
Query: 459 QPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPG 518
+P + ++L RPWK YSR +FM+++I D+I P G+ W+G L+T + EY GPG
Sbjct: 2 EPVKYSFPTYLGRPWKEYSRTVFMQSSISDVIDPAGWHEWSGNFALNTLVYREYQYPGPG 181
Query: 519 SNVQARVKWG--KGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
+ RV W K + S ++A +T +I G WL +TG PF LG
Sbjct: 182 AGTSKRVAWKGVKVITSASEAQAFTPGSFIAGSNWLGSTGFPFSLG 319
>BF177669
Length = 336
Score = 89.4 bits (220), Expect = 1e-18
Identities = 42/99 (42%), Positives = 61/99 (61%), Gaps = 1/99 (1%)
Frame = +3
Query: 444 GVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQF 503
G+ +QNC I P L V+++L RPWK YS + M++T+G I P+G+LPW G
Sbjct: 42 GISIQNCSISPFGNLS----YVQTYLGRPWKNYSTTVVMQSTLGSFISPNGWLPWVGNSA 209
Query: 504 LDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYT 541
DT F+AEY N G G++ + RVKW G ++ A+K+T
Sbjct: 210 PDTIFYAEYQNVGQGASTKNRVKWKGLKTITSKQASKFT 326
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,968,519
Number of Sequences: 28460
Number of extensions: 92231
Number of successful extensions: 416
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 386
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 388
length of query: 566
length of database: 4,897,600
effective HSP length: 95
effective length of query: 471
effective length of database: 2,193,900
effective search space: 1033326900
effective search space used: 1033326900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144728.4


