

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.1 - phase: 0
(999 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
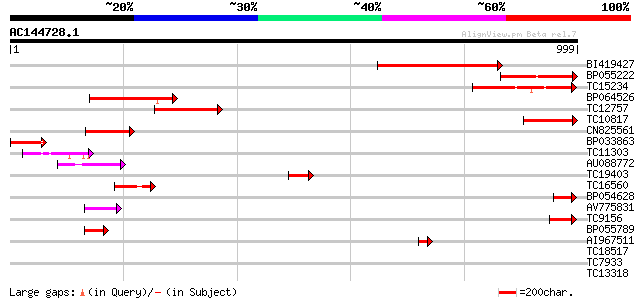
Score E
Sequences producing significant alignments: (bits) Value
BI419427 375 e-104
BP055222 228 4e-60
TC15234 similar to PIR|T47827|T47827 squamosa promoter binding p... 200 8e-52
BP064526 199 1e-51
TC12757 174 5e-44
TC10817 similar to UP|O80717 (O80717) At2g47080 protein, partial... 143 1e-34
CN825561 122 2e-28
BP033863 100 1e-21
TC11303 homologue to UP|O82651 (O82651) Squamosa-promoter bindin... 91 1e-18
AU088772 90 2e-18
TC19403 similar to UP|O82651 (O82651) Squamosa-promoter binding ... 84 8e-17
TC16560 similar to UP|Q9S849 (Q9S849) Squamosa promoter binding ... 76 2e-14
BP054628 70 1e-12
AV775831 62 6e-10
TC9156 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Frag... 50 1e-06
BP055789 47 1e-05
AI967511 47 1e-05
TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial ... 37 0.016
TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonas... 33 0.29
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 30 1.9
>BI419427
Length = 661
Score = 375 bits (963), Expect = e-104
Identities = 186/220 (84%), Positives = 200/220 (90%)
Frame = +1
Query: 649 SFFPFIVAEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQL 708
SFFPFIVAEEDVC++I LEPLLE SETDPDIEGTGKIKAKSQAMDFIHE+GWLLH SQL
Sbjct: 1 SFFPFIVAEEDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQL 180
Query: 709 KYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALS 768
K RMV+L+S DLFPL+RF WLMEFSMDHDWCAVVKKLLNLLL+ TV+ GDHP+L ALS
Sbjct: 181 KSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDLALS 360
Query: 769 EMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLT 828
EMGLLHRAVRRNSKQLVELLL YVP+N S+E+ PE KALV G+ S+LFRPDA GPAGLT
Sbjct: 361 EMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLT 540
Query: 829 PLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGST 868
PLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGST
Sbjct: 541 PLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGST 660
>BP055222
Length = 536
Score = 228 bits (581), Expect = 4e-60
Identities = 111/135 (82%), Positives = 121/135 (89%)
Frame = -1
Query: 865 TGSTPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEI 924
TGSTPEDYARLRGHYTYIHLVQKKINK QGA HVVVEIP+N+TE++ N KQNES T+ EI
Sbjct: 536 TGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEI 357
Query: 925 GKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLY 984
K R QG+CKLCD K+SCRTAVGRS+VY+PAMLSMVA+AAVCVCVALLFKSSPEVLY
Sbjct: 356 AKT---RDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLY 186
Query: 985 MFRPFRWESLDFGTS 999
MFRPF WESLDFGTS
Sbjct: 185 MFRPFSWESLDFGTS 141
>TC15234 similar to PIR|T47827|T47827 squamosa promoter binding protein-like
12 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (11%)
Length = 737
Score = 200 bits (509), Expect = 8e-52
Identities = 108/189 (57%), Positives = 130/189 (68%), Gaps = 5/189 (2%)
Frame = +2
Query: 815 YLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYAR 874
+LFRPDAVGP GLTPLH+AA GSE+VLDALT+DP MVGIEAW++ARD+TG TP DYA
Sbjct: 17 FLFRPDAVGPGGLTPLHVAASMSGSENVLDALTDDPGMVGIEAWESARDNTGLTPNDYAS 196
Query: 875 LRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNE-----SFTSLEIGKAEV 929
LRGHYTYI LVQKK +K +G V++IP + +S KQ+E +SL+ K E
Sbjct: 197 LRGHYTYIQLVQKKTSK-KGGRQCVLDIPGTLVDSYTKQKQSERHGTSKVSSLQTVKIET 373
Query: 930 RRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPF 989
C LC K + ++VYRP MLSMV IAAVCVCVAL FKS P V YM +PF
Sbjct: 374 TTVPLQCGLCKHK-QAYGGMRPALVYRPVMLSMVTIAAVCVCVALFFKSLPRV-YMTQPF 547
Query: 990 RWESLDFGT 998
WESL +G+
Sbjct: 548 NWESLKYGS 574
>BP064526
Length = 533
Score = 199 bits (507), Expect = 1e-51
Identities = 100/160 (62%), Positives = 122/160 (75%), Gaps = 5/160 (3%)
Frame = +1
Query: 141 TSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEF 200
T RAVCQV+DC ADLS KDYHRRHKVCE HSKA+ ALVGN MQRFCQQCSRFH+L+EF
Sbjct: 34 TLKRAVCQVQDCRADLSSAKDYHRRHKVCEGHSKATMALVGNVMQRFCQQCSRFHVLQEF 213
Query: 201 DEGKRSCRRRLAGHN-KRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMHYQPTD- 258
DEGKRSCRRRLAGHN +RRRKT+ +A NG ND++ SSYLL SL++ILSNMH +D
Sbjct: 214 DEGKRSCRRRLAGHNRRRRRKTHPDAEVNGEALNDERGSSYLLTSLIRILSNMHSNSSDH 393
Query: 259 ---QDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREG 295
D+L+HLLR+LAS ++L+++L + L+ G
Sbjct: 394 TRNNDILSHLLRNLASLAGTINGRSLASILEGSQGLVNAG 513
>TC12757
Length = 710
Score = 174 bits (442), Expect = 5e-44
Identities = 90/121 (74%), Positives = 99/121 (81%), Gaps = 1/121 (0%)
Frame = +3
Query: 255 QPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQG 314
Q TDQDLLTHLLRSLAS+N EQG NLSN+ RE ENLLREGGSSR S MV L SNGSQG
Sbjct: 348 QTTDQDLLTHLLRSLASRNSEQGGNNLSNIYREPENLLREGGSSRESEMVPTLLSNGSQG 527
Query: 315 SPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDS-SGQTK 373
SPT I QHQ VSMN+MQQE+VH HD R +D QL+S IKPSISN+PPAY+E RDS +GQ K
Sbjct: 528 SPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPPAYAEARDSTAGQIK 707
Query: 374 M 374
M
Sbjct: 708 M 710
>TC10817 similar to UP|O80717 (O80717) At2g47080 protein, partial (13%)
Length = 648
Score = 143 bits (361), Expect = 1e-34
Identities = 71/95 (74%), Positives = 80/95 (83%), Gaps = 1/95 (1%)
Frame = +2
Query: 906 MTESNKNPKQNESF-TSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVA 964
+T N N Q+E TS EIGKAEV+ Q CKLCD K+SCRTA GRS+VY+PAMLSMVA
Sbjct: 2 LTRFNANEXQDELLSTSFEIGKAEVKSVQKLCKLCDLKLSCRTAAGRSLVYKPAMLSMVA 181
Query: 965 IAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
+AAVCVCVALLFKSSPEVLY+FRPFRWESL+FGTS
Sbjct: 182 VAAVCVCVALLFKSSPEVLYIFRPFRWESLEFGTS 286
>CN825561
Length = 646
Score = 122 bits (307), Expect = 2e-28
Identities = 57/87 (65%), Positives = 63/87 (71%)
Frame = +2
Query: 134 KSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSR 193
K GG+S CQVE+C ADLS K YHRRHKVCE H+KA + QRFCQQCSR
Sbjct: 170 KRGSKGGSSVPPSCQVENCDADLSEAKQYHRRHKVCEYHAKAHAVHIAGMHQRFCQQCSR 349
Query: 194 FHILEEFDEGKRSCRRRLAGHNKRRRK 220
FH L EFD+ KRSCRRRLAGHN+RRRK
Sbjct: 350 FHELSEFDDSKRSCRRRLAGHNERRRK 430
>BP033863
Length = 422
Score = 100 bits (249), Expect = 1e-21
Identities = 47/65 (72%), Positives = 52/65 (79%)
Frame = +2
Query: 1 MGERLGAENYHFYGVGGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQ 60
M R GAE YH+YGVG SSD+ GMGKRS EW+LN+WRWDGDLFIASRVN VQ + VGQ
Sbjct: 233 MEARFGAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDG--VGQ 406
Query: 61 QFFPL 65
Q FPL
Sbjct: 407 QLFPL 421
>TC11303 homologue to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (3%)
Length = 512
Score = 90.5 bits (223), Expect = 1e-18
Identities = 59/138 (42%), Positives = 78/138 (55%), Gaps = 13/138 (9%)
Frame = -3
Query: 23 GMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSC 82
G GKRS EW+LNDW+WDGDLF A +N V ++ G QFFP P ++N SS+
Sbjct: 402 GNGKRSLEWDLNDWKWDGDLFTAQPLNSVPSDCR--GGQFFPPAPEFP--AKTANLSSNL 235
Query: 83 SEEGDLEKGNKEGEKKRRVIVL----EDDGLNDKAGALSLNLAGHVSPVV----ERDGKK 134
S + +G +E EK+RR + + E D ND G+L+LNL G V PV+ E+ GKK
Sbjct: 234 SASVNSGEGKRELEKRRRGVTVRGEGEGDEGNDGNGSLNLNLGGQVYPVMVEGEEKIGKK 55
Query: 135 SR-----GAGGTSNRAVC 147
+R T RAVC
Sbjct: 54 TRVIETTAYATTFKRAVC 1
>AU088772
Length = 422
Score = 90.1 bits (222), Expect = 2e-18
Identities = 47/121 (38%), Positives = 64/121 (52%)
Frame = +1
Query: 84 EEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKKSRGAGGTSN 143
EE + E+ ++ E + ++ +DG+ K V + K+ AGG++
Sbjct: 64 EEDECEQXQEDXEXEEEAVLYGEDGIRKKRVV-----------VTDLYSKRGSKAGGSAV 210
Query: 144 RAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEG 203
CQVE C ADLS K HRRHKVCE H+KA L+ QRFC QC RFH L +FD+
Sbjct: 211 PPSCQVEGCYADLSGAKAXHRRHKVCEHHAKAPSXLLSEQQQRFCXQCXRFHELSQFDDP 390
Query: 204 K 204
K
Sbjct: 391 K 393
>TC19403 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (5%)
Length = 499
Score = 84.3 bits (207), Expect = 8e-17
Identities = 38/44 (86%), Positives = 42/44 (95%)
Frame = +3
Query: 491 VLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIR 534
VLTIYLRQAEAVWEE+C DLTSSL +LLDVSDDTFW+TGWVHI+
Sbjct: 3 VLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIQ 134
>TC16560 similar to UP|Q9S849 (Q9S849) Squamosa promoter binding
protein-like 8, partial (18%)
Length = 622
Score = 76.3 bits (186), Expect = 2e-14
Identities = 38/73 (52%), Positives = 49/73 (67%)
Frame = +2
Query: 185 QRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLIS 244
QRFCQQCSRFH+L EFD GKRSCR+RLA HN+RRRKT Q T + Q S+ L +
Sbjct: 29 QRFCQQCSRFHLLSEFDNGKRSCRKRLADHNRRRRKTQQS-------TQEIQKSTTALDN 187
Query: 245 LLKILSNMHYQPT 257
+ + ++ QP+
Sbjct: 188 IARSPADSGTQPS 226
>BP054628
Length = 508
Score = 70.5 bits (171), Expect = 1e-12
Identities = 32/40 (80%), Positives = 36/40 (90%)
Frame = -2
Query: 959 MLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
MLSMVAIAAVCVCVALLFKSSP V Y+F PF WESL++G+
Sbjct: 507 MLSMVAIAAVCVCVALLFKSSPRVYYVFHPFSWESLEYGS 388
>AV775831
Length = 475
Score = 61.6 bits (148), Expect = 6e-10
Identities = 28/66 (42%), Positives = 38/66 (57%), Gaps = 2/66 (3%)
Frame = -3
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
K++R + A C V+ C +DLS +DYHRRHKVCE+HS+ + Q C CS
Sbjct: 449 KRARAVHYATLTAFCLVDGCSSDLSNCRDYHRRHKVCELHSQTPEVTIAGMKQGLCLPCS 270
Query: 193 R--FHI 196
R FH+
Sbjct: 269 RSSFHM 252
>TC9156 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fragment),
partial (3%)
Length = 850
Score = 50.4 bits (119), Expect = 1e-06
Identities = 23/48 (47%), Positives = 35/48 (72%)
Frame = +2
Query: 951 RSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
+ ++RP + S++A+AAVCVCV + F+ +P V PFRWE+LD+GT
Sbjct: 2 KGWLHRPFIHSILAVAAVCVCVCVFFRGTPFV-GSEGPFRWENLDYGT 142
>BP055789
Length = 455
Score = 47.4 bits (111), Expect = 1e-05
Identities = 22/41 (53%), Positives = 29/41 (70%)
Frame = +3
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHS 173
K+SR G+ N A C V+ C +DLS ++YHRRH+VCE HS
Sbjct: 336 KRSRLHNGSLNMA-CSVDGCTSDLSDCREYHRRHRVCEKHS 455
>AI967511
Length = 375
Score = 47.0 bits (110), Expect = 1e-05
Identities = 20/25 (80%), Positives = 20/25 (80%)
Frame = +2
Query: 721 LFPLQRFTWLMEFSMDHDWCAVVKK 745
LFPL RF WLMEF MD DWCAV KK
Sbjct: 62 LFPLIRFKWLMEFPMDCDWCAVAKK 136
>TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (4%)
Length = 493
Score = 37.0 bits (84), Expect = 0.016
Identities = 21/47 (44%), Positives = 27/47 (56%)
Frame = +2
Query: 399 STNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQS 445
ST ATSS PW+ S SS + +SG +S+ SPSSS G + S
Sbjct: 242 STTSATSSATTPWS---SSSSSSSPSSGTPSPSSSSSPSSSPGSSSS 373
>TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonase
inhibiting protein, partial (78%)
Length = 1320
Score = 32.7 bits (73), Expect = 0.29
Identities = 30/91 (32%), Positives = 46/91 (49%), Gaps = 4/91 (4%)
Frame = +1
Query: 355 ISNSPPAYSETRDSS----GQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYP 410
+SN+ P +E+R SS QT+++ + ++V S L ST+ +TS +P
Sbjct: 184 VSNATPKLTESRHSSYPPLSQTQISQAQYHLLWVTS---------LTSSTSTSTS---FP 327
Query: 411 WTQQDSHQSSPAQTSGNSDSASAQSPSSSSG 441
SSPAQ+S S S+ + SPS S G
Sbjct: 328 --------SSPAQSSPPSPSSPSSSPSPSPG 396
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 30.0 bits (66), Expect = 1.9
Identities = 17/54 (31%), Positives = 29/54 (53%)
Frame = -3
Query: 397 PVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRI 450
P + +TSS+ + S SS + +S +S S+S+ S SSSS + S + +
Sbjct: 409 PSFPSTSTSSLPSSSSSSSSSSSSSSPSSSSSSSSSSSSSSSSSSSSSSSSSSL 248
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,219,333
Number of Sequences: 28460
Number of extensions: 241749
Number of successful extensions: 1470
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1434
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1456
length of query: 999
length of database: 4,897,600
effective HSP length: 99
effective length of query: 900
effective length of database: 2,080,060
effective search space: 1872054000
effective search space used: 1872054000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144728.1


