

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.4 - phase: 0
(762 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
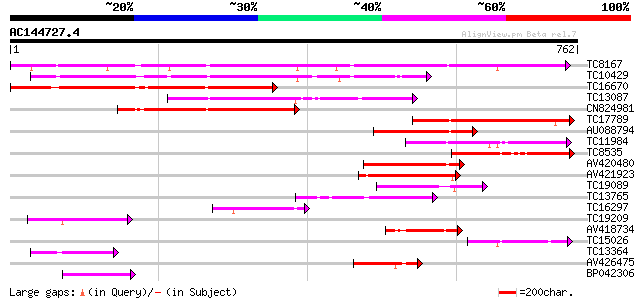
Score E
Sequences producing significant alignments: (bits) Value
TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine prote... 501 e-142
TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%) 338 1e-93
TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 314 3e-86
TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like prote... 235 2e-62
CN824981 231 3e-61
TC17789 similar to PIR|T05768|T05768 subtilisin-like proteinase ... 194 3e-50
AU088794 149 2e-36
TC11984 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, ... 148 3e-36
TC8535 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like... 136 1e-32
AV420480 134 5e-32
AV421923 127 9e-30
TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C... 123 1e-28
TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine end... 106 2e-23
TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type pro... 99 3e-21
TC19209 similar to GB|BAB11244.1|10177874|AB010074 serine protea... 97 1e-20
AV418734 95 5e-20
TC15026 weakly similar to UP|P93205 (P93205) Serine protease, SB... 89 3e-18
TC13364 similar to UP|Q9LUM3 (Q9LUM3) Subtilisin proteinase-like... 87 8e-18
AV426475 85 4e-17
BP042306 82 4e-16
>TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (65%)
Length = 2760
Score = 501 bits (1290), Expect = e-142
Identities = 298/785 (37%), Positives = 443/785 (55%), Gaps = 32/785 (4%)
Frame = +3
Query: 1 MASNICLWLWFSYITSLHVIFTLALSD---NYIIHMNLSDMPKSFSNQHSWYESTLAQVT 57
MAS L F++I L + + S YI+HMN P+ + + WY ++L ++
Sbjct: 57 MASIPISTLSFTFILLLLQCLSASCSSPKKTYIVHMNHHTKPQIYPTRRDWYTASLRSLS 236
Query: 58 TTNNNLNNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSP 117
T ++ ++ + Y Y NGF+A+L ++ ++L + D L TT +P
Sbjct: 237 LTTDS--ETSDDPLLYAYDTAYNGFAASLDEQQAQTLLGSDSVLGLYEDTLYHLHTTRTP 410
Query: 118 QFLGLNPYRGAWP------TSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLC 171
QFLGL+ G W +D+I+GV+DTGVWPES SF D GM +IPS+W+G+ C
Sbjct: 411 QFLGLDTQTGLWEGHRTLELDQASRDVIIGVLDTGVWPESPSFNDAGMPEIPSRWRGE-C 587
Query: 172 QFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSNISTTILN----STRDTNGHGTHTST 227
+ N + SLCN+KLIGAR F++GF N S RD++GHGTHT++
Sbjct: 588 E----NATDFSSSLCNRKLIGARSFSRGFHMAAGNDGGFGKEREPPSPRDSDGHGTHTAS 755
Query: 228 TAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVD 287
TAAGS V AS GYA+GTARG+A +RVA YK W DG +SDI+A +D AI DGVD
Sbjct: 756 TAAGSHVGNASLLGYASGTARGMAPQARVATYKVCWS-DG-CFASDILAGMDRAIRDGVD 929
Query: 288 ILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGT 347
+LS+SLG ++D +AI FAAME+GIFVS SAGN+GPS S+ N PW++TV AGT
Sbjct: 930 VLSLSLGGGSGPYFRDTIAIGAFAAMERGIFVSCSAGNSGPSKASLANVAPWLMTVGAGT 1109
Query: 348 LDREFLGTVTLGNGVSLTGLSFYLG-NFSANNFPIVFMG-------MC-DNVKELNTVKR 398
LDR+F + LGN G+S Y G A +V+ +C + V+
Sbjct: 1110LDRDFPASALLGNKKRFAGVSLYSGKGMGAEPVGLVYSKGSNQSGILCLPGSLDPAVVRG 1289
Query: 399 KIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDV---DNSFPSIIINPVNGEIVK 455
K+V+C+ E+ V +A +G + + + ++ + P++ + + G+ ++
Sbjct: 1290KVVLCDRGLNARVEKGKVVKEAGGIGMILTNTAANGEELVADSHLLPAVAVGRIVGDQIR 1469
Query: 456 AYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILA 515
Y+ S+ + A +SF T V+ +P V +SSRGP+ +LKPD+ PG +ILA
Sbjct: 1470EYV---TSDPNPTAVLSFSGTVLNVRPSPVVAAFSSRGPNMITKQILKPDVIGPGVNILA 1640
Query: 516 AWPTNVPVSNFGTEV-FNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTS 574
W + S + + FN++ GTSMSCPH++G+ ALLK AH WSPS+I+SA+MTT+
Sbjct: 1641GWSEAIGPSGLPQDSRKSQFNIMSGTSMSCPHISGLGALLKAAHPDWSPSAIKSALMTTA 1820
Query: 575 DILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQ 634
+ DNT ++D G +TP+A GAGH+NP +AL PGLVYD +DYI LC+L+++
Sbjct: 1821YVHDNTNSPLRDAA-GGEFSTPWAHGAGHVNPQKALSPGLVYDAKARDYIAFLCSLDYSP 1997
Query: 635 KNISAITRSSFNDCSKPSLD---LNYPSF-IAFSNARNSSRTTNEFHRTVTNVGEKKTTY 690
++ I + + +CS+ D LNYPSF + F + ++ + RTVTNVGE + Y
Sbjct: 1998DHLQLIVKRAGVNCSRKLSDPGQLNYPSFSVVFDSVVFDAKKVVRYTRTVTNVGEAGSVY 2177
Query: 691 FASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKV--AFGYLSWRDGKHV 748
+ +TV P KL F K E+ Y + + + V AFG ++W++ +H
Sbjct: 2178DVVVDGPSMVGITVYPTKLEFGKVGERKRYTVTFVSKKGASDDLVRSAFGSITWKNEQHQ 2357
Query: 749 VRSPI 753
VRSP+
Sbjct: 2358VRSPV 2372
>TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%)
Length = 1742
Score = 338 bits (868), Expect = 1e-93
Identities = 215/561 (38%), Positives = 308/561 (54%), Gaps = 22/561 (3%)
Frame = +3
Query: 29 YIIHMNLSD--MPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANL 86
YIIH+ + M + SWY S L ++ ++ Y+Y NV+ GF+A+L
Sbjct: 153 YIIHVTGPEGKMLTESEDLESWYHSFLPPTLKSSEE-----QPRVIYSYKNVLRGFAASL 317
Query: 87 SPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTG 146
+ EE +++ I + P+FLGL G W S+FGK +I+GV+D+G
Sbjct: 318 TQEELSAVERKMALFQLILRGCYIVKPLIPPKFLGLQQDTGVWKESNFGKGVIIGVLDSG 497
Query: 147 VWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSN 206
+ P SF D G+ P KWKG+ +N++ CN KLIGAR FN A +
Sbjct: 498 ITPGHPSFSDVGIPPPPPKWKGRC---------DLNVTACNNKLIGARAFNLAAEAMNGK 650
Query: 207 ISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKD 266
+ + D +GHGTHT++TAAG+ V+ A G A GTA G+A + +AIYK +G+D
Sbjct: 651 KAEAPI----DEDGHGTHTASTAAGAFVNYAEVLGNAKGTAAGMAPHAHLAIYKVCFGED 818
Query: 267 GDALSSDIIAAIDAAISDGVDILSISLG-SDDLLLYKDPVAIATFAAMEKGIFVSTSAGN 325
SDI+AA+DAA+ DGVD++SISLG S+ + D AI FAAM+KGIFVS +AGN
Sbjct: 819 --CPESDILAALDAAVEDGVDVISISLGLSEPPPFFNDSTAIGAFAAMQKGIFVSCAAGN 992
Query: 326 NGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLS-FYLGNFSANNFPIVFM 384
+GP SI N PW++TV A T+DR + T LGNG G S F +F+ P+ +
Sbjct: 993 SGPFNSSIVNAAPWILTVGASTIDRRIVATAKLGNGQEFDGESVFQPSSFTPTLLPLAYA 1172
Query: 385 G--------MCDNVK-ELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDIN 435
G C N + + + K+V+CE + + K + V + ++ +N
Sbjct: 1173GKNGKEESAFCANGSLDDSAFRGKVVLCERGG-----GIARIAKGEEVKRAGGAAMILMN 1337
Query: 436 DVDNSF---------PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSV 486
D N+F P+ ++ G +KAYI NS A+ A + FK T G P+V
Sbjct: 1338DETNAFSLSADVHALPATHVSYAAGIEIKAYI---NSTATPTATILFKGTVIGNSLAPAV 1508
Query: 487 DFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPH 546
+SSRGP+ P +LKPDI PG +ILAAWP P+SN T+ FN+ GTSMSCPH
Sbjct: 1509ASFSSRGPNLPSPGILKPDIIGPGVNILAAWP--FPLSN-STDSKLTFNIESGTSMSCPH 1679
Query: 547 VAGVAALLKGAHNGWSPSSIR 567
++G+AALLK +H WSP++I+
Sbjct: 1680LSGIAALLKSSHPHWSPAAIK 1742
>TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (42%)
Length = 1183
Score = 314 bits (805), Expect = 3e-86
Identities = 177/361 (49%), Positives = 231/361 (63%), Gaps = 1/361 (0%)
Frame = +2
Query: 1 MASNICLWLWFSYITSLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTN 60
MAS L ++++L + T YI+H+ S+MP+SF + +WYES+L V
Sbjct: 149 MASFHTSTLLVLFLSTLCIHATPENKGTYIVHVAKSEMPESFEHHAAWYESSLQSV---- 316
Query: 61 NNLNNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFL 120
S S+++ YTY N ++GFS L+PEE L+ +G ++ +P+ +L TT +P FL
Sbjct: 317 -----SDSAEMLYTYDNAIHGFSTRLTPEEARLLEAQTGVLAVLPERKFELHTTRTPLFL 481
Query: 121 GLNPYRGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQS 180
GL+ +P S G ++IVGV+DTGVWPES+SF D G +PS WKG+ C+ +N +
Sbjct: 482 GLDKSADMFPASGAGGEVIVGVLDTGVWPESKSFDDTGFGPVPSTWKGE-CE-SGTNFTA 655
Query: 181 INLSLCNKKLIGARFFNKGFLAKHSNISTTILN-STRDTNGHGTHTSTTAAGSKVDGASF 239
N CNKKLIGAR+F+KG A I T + S RD +GHGTHT++TAAGS V GAS
Sbjct: 656 AN---CNKKLIGARYFSKGAEAMLGPIDETAESKSPRDDDGHGTHTASTAAGSVVSGASL 826
Query: 240 FGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLL 299
FGYA GTARG+A +RVA YK W G SSDI+AAID AI+D V++LS+SLG
Sbjct: 827 FGYAAGTARGMAPHARVAAYKVCW--KGGCFSSDILAAIDKAIADNVNVLSLSLGGGMAD 1000
Query: 300 LYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLG 359
Y+D VAI FAAMEKGI VS SAGN GPS S+ N PW+ TV AGTLDR+F V LG
Sbjct: 1001YYRDSVAIGAFAAMEKGILVSCSAGNAGPSAYSLSNVAPWITTVGAGTLDRDFPALVELG 1180
Query: 360 N 360
N
Sbjct: 1181N 1183
>TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like protein, partial
(31%)
Length = 1036
Score = 235 bits (599), Expect = 2e-62
Identities = 143/351 (40%), Positives = 204/351 (57%), Gaps = 15/351 (4%)
Frame = +2
Query: 213 NSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSS 272
N+ RD GHG+HT++TAAG+ V GASF+G A GTARG S+R+A YK + G +
Sbjct: 17 NTVRDIEGHGSHTASTAAGNTVIGASFYGLAQGTARGAVPSARIAAYKVCYPNHG-CDDA 193
Query: 273 DIIAAIDAAISDGVDILSISLGSDDLL-LYKDPVAIATFAAMEKGIFVSTSAGNNGPSFK 331
+I+AA D AI+DGV+ILS+SLG++ + L D VAI +F AME+GI +AGN+GP+
Sbjct: 194 NILAAFDDAIADGVNILSVSLGTNYCVDLLSDTVAIGSFHAMERGILTLQAAGNSGPASP 373
Query: 332 SIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFSANNFPIV--------- 382
SI + PW++TVAA T+DR+F+ V LGNG++L G S + FPI
Sbjct: 374 SICSVAPWLLTVAASTIDRQFIDKVLLGNGMTLIGRSVNAVVSNGTKFPIAKWNASDERC 553
Query: 383 --FMGMCDNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVD-- 438
F +C + +VK K+V+CEG +H++ Y VG + NDV
Sbjct: 554 KGFSDLCQCLGS-ESVKGKVVLCEGR---MHDEA--AYLDGAVGSITEDLTNPDNDVSFV 715
Query: 439 NSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSC 498
FPS+ +NP + IV+ Y S + + I K F +S P V +SSRGP+
Sbjct: 716 TYFPSVNLNPKDYAIVQNYTNSAKNPKAEI----LKSEIFKDRSAPKVVSFSSRGPNPLI 883
Query: 499 PYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFN-NFNLIDGTSMSCPHVA 548
P ++KPD++APG ILAAW S + ++ +N++ GTSM+CPHVA
Sbjct: 884 PEIMKPDVSAPGVDILAAWSPEAAPSEYSSDKRKVRYNVVSGTSMACPHVA 1036
>CN824981
Length = 726
Score = 231 bits (589), Expect = 3e-61
Identities = 126/246 (51%), Positives = 160/246 (64%), Gaps = 2/246 (0%)
Frame = +1
Query: 146 GVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHS 205
GVWPE +S D G++ +PS WKGQ C+ N+ +N S CN+KLIGARFF+KG+ A
Sbjct: 4 GVWPELKSLDDTGLSPVPSTWKGQ-CEAGNN----MNSSSCNRKLIGARFFSKGYEATLG 168
Query: 206 NIS-TTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWG 264
I +T S RD +GHG+HT TTAAGS V GAS FG A+GTARG+A+ +RVA YK W
Sbjct: 169 PIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWL 348
Query: 265 KDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAG 324
G SSDI A ID AI DGV+I+S+S+G ++D +AI F A GI VSTSAG
Sbjct: 349 --GGCFSSDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAG 522
Query: 325 NNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGN-FSANNFPIVF 383
N GPS S+ N PW+ TV AGT+DR+F +TLGN ++ TG S Y G S + P+V+
Sbjct: 523 NGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVY 702
Query: 384 MGMCDN 389
G N
Sbjct: 703 AGNASN 720
>TC17789 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (14%)
Length = 940
Score = 194 bits (494), Expect = 3e-50
Identities = 106/223 (47%), Positives = 143/223 (63%), Gaps = 6/223 (2%)
Frame = +2
Query: 542 MSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGA 601
M+CPH++G AALLK AH WSP+++RSA+MTT+ +LDN K+ + D GN ++TP+ GA
Sbjct: 2 MACPHISGAAALLKSAHPDWSPAAVRSAMMTTATVLDNRKQILTDEATGN-SSTPYDFGA 178
Query: 602 GHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFN-DCSKPSLD-LNYPS 659
GH+N A+DPGLVYDI DY+N LC + + + I ITR+ + KPS + LNYPS
Sbjct: 179 GHVNLGLAMDPGLVYDITGADYVNFLCGIGYGPRVIQVITRAPVSCPARKPSPENLNYPS 358
Query: 660 FIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASI-TPIKGFRVTVIPNKLVFKKKNEKI 718
F+A + TT F RTVTNVG + Y S+ + +KG VTV P++LVF + +K
Sbjct: 359 FVAMFPVGSKGLTTKTFIRTVTNVGPANSVYRVSVESQMKGVTVTVKPSRLVFSEAVKKR 538
Query: 719 SYKLKIEGPRMTQK---NKVAFGYLSWRDGKHVVRSPIVVTNI 758
SY + + K + FG L+W DGKHVVRSPIVVT I
Sbjct: 539 SYVVTVAAETRFLKMGPSGAVFGSLTWTDGKHVVRSPIVVTQI 667
>AU088794
Length = 433
Score = 149 bits (375), Expect = 2e-36
Identities = 70/141 (49%), Positives = 99/141 (69%), Gaps = 1/141 (0%)
Frame = +2
Query: 489 YSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNN-FNLIDGTSMSCPHV 547
+S+RGP+ P +LKPD+ PG +ILAAWP S ++V FN++ GTSM+CPHV
Sbjct: 14 FSARGPNPESPEILKPDVIXPGLNILAAWPDXXGPSGVPSDVRRTEFNILSGTSMACPHV 193
Query: 548 AGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPN 607
+G+AALLK AH WSP++I+SA+MTT+ +DN + + D NGN + F G+GH++P
Sbjct: 194 SGLAALLKAAHPXWSPAAIKSALMTTAYTVDNKGDAMLDESNGN-VSLVFDYGSGHVHPX 370
Query: 608 RALDPGLVYDIGVQDYINLLC 628
+A+DPGLVYDI DY++ LC
Sbjct: 371 KAMDPGLVYDISTYDYVDFLC 433
>TC11984 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, partial
(30%)
Length = 966
Score = 148 bits (374), Expect = 3e-36
Identities = 88/232 (37%), Positives = 129/232 (54%), Gaps = 8/232 (3%)
Frame = +1
Query: 532 NNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGN 591
+ +N+I GTSMSCPHV+G+A +K H WS S+I+SAIMT++ +N K + +
Sbjct: 52 SQYNIISGTSMSCPHVSGLAGSIKSRHTTWSASAIKSAIMTSATQNNNLKAPMTT--DSG 225
Query: 592 RAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRS---SFNDC 648
ATP+ GAG + + + PGLVY+ DY+N LC + I I+R+ SF
Sbjct: 226 SVATPYDYGAGEMTTSASFQPGLVYETSTTDYLNYLCYIGLNVTTIKVISRTVPDSFTCP 405
Query: 649 SKPSLD----LNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPI-KGFRVT 703
S D +NYPS IA +N + + RTVTNVGE+ T ++ I G +
Sbjct: 406 QDSSADHVSNINYPS-IAITNF--NGKGAINVSRTVTNVGEEDETVYSPIVDAPSGVNIK 576
Query: 704 VIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVV 755
+IP KL F K ++K+SY + T + FG L+W +GK++VRSP V+
Sbjct: 577 LIPEKLPFTKSSQKLSYPVIFSLTSSTSLKEDLFGSLAWSNGKYIVRSPFVL 732
>TC8535 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein,
partial (10%)
Length = 780
Score = 136 bits (343), Expect = 1e-32
Identities = 74/166 (44%), Positives = 104/166 (62%), Gaps = 2/166 (1%)
Frame = +2
Query: 595 TPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFNDC--SKPS 652
TPFA G+GH+ PN A+DPGLVYD+ + DY+N LCA + Q+ ISA+ + C S
Sbjct: 2 TPFAYGSGHVQPNSAIDPGLVYDLSIVDYLNFLCASGYDQQLISALNFNKTFTCSGSHSI 181
Query: 653 LDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFK 712
DLNYPS I N R++ RTVTNVG + YFA+ + G ++ V+PN L FK
Sbjct: 182 TDLNYPS-ITLPNIRSNVVNVT---RTVTNVG-PPSKYFAN-AQLPGHKIVVVPNSLSFK 343
Query: 713 KKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVTNI 758
K EK ++++ ++ +TQ+ K FG L W +G+H+VRSPI V +I
Sbjct: 344 KIGEKKTFQVIVQATSVTQRRKYLFGKLQWTNGRHIVRSPITVRHI 481
>AV420480
Length = 412
Score = 134 bits (337), Expect = 5e-32
Identities = 66/137 (48%), Positives = 97/137 (70%), Gaps = 1/137 (0%)
Frame = +3
Query: 476 TAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFN-NF 534
T G P+V +S+RGPS + P +LKPD+ APG +I+AAWP N+ ++ ++ NF
Sbjct: 9 TVIGNSRAPAVATFSARGPSFTNPSILKPDVVAPGVNIIAAWPQNLGPTSLPQDLRRVNF 188
Query: 535 NLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
+++ GTSMSCPHV+G+AAL+ AH WSP++I+SAIMTT+D+ D+ K I D ++ A
Sbjct: 189 SVMSGTSMSCPHVSGIAALVHSAHPKWSPAAIKSAIMTTADVTDHMKRPILD---EDKPA 359
Query: 595 TPFALGAGHINPNRALD 611
FA+GAG++NP RAL+
Sbjct: 360 GVFAIGAGNVNPQRALN 410
>AV421923
Length = 463
Score = 127 bits (318), Expect = 9e-30
Identities = 71/149 (47%), Positives = 91/149 (60%), Gaps = 11/149 (7%)
Frame = +2
Query: 469 ANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGT 528
A++SF T FG P V +SSRGPS V+KPD+TAPG ILAAWP+ S
Sbjct: 14 ASISFIGTRFG-DPAPVVAAFSSRGPSIVGLDVIKPDVTAPGVHILAAWPSKTSPSMLKN 190
Query: 529 EVFNN-FNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDI 587
+ FN+I GTSMSCPHV+GVAAL++ H WSP++++SA+MTT+ L+N I DI
Sbjct: 191 DKRRVLFNIISGTSMSCPHVSGVAALIRSVHRDWSPAAVKSALMTTAYTLNNKGAPISDI 370
Query: 588 GNGNRA----------ATPFALGAGHINP 606
G N A PFA G+GH+NP
Sbjct: 371 GASNSISNSNSNHSGFAVPFAFGSGHVNP 457
>TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, partial
(16%)
Length = 600
Score = 123 bits (309), Expect = 1e-28
Identities = 68/154 (44%), Positives = 89/154 (57%), Gaps = 4/154 (2%)
Frame = +1
Query: 493 GPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNF-GTEVFNNFNLIDGTSMSCPHVAGVA 551
GP+ P +LKPDI APG ++LAAW P+S G + +N I GTSM+CPH A A
Sbjct: 16 GPNPITPNILKPDIAAPGVTVLAAWSPLNPISEVEGDKRKLPYNAISGTSMACPHAAAAA 195
Query: 552 ALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATP---FALGAGHINPNR 608
A +K H WSP+ I+SA+MTT+ + N A P FA GAG INP +
Sbjct: 196 AYVKSFHPNWSPAMIKSALMTTATPM-------------NSALNPEAEFAYGAGQINPVK 336
Query: 609 ALDPGLVYDIGVQDYINLLCALNFTQKNISAITR 642
A++PGLVYDI QDYI LC +T + +T+
Sbjct: 337 AVNPGLVYDITEQDYIQFLCGXGYTDTLLQNLTQ 438
>TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine endopeptidase
XSP1, partial (20%)
Length = 635
Score = 106 bits (264), Expect = 2e-23
Identities = 75/194 (38%), Positives = 105/194 (53%), Gaps = 4/194 (2%)
Frame = +1
Query: 385 GMC-DNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDNSF-- 441
G C ++ + N VK K+V C+ N V K +G + S+ D+ F
Sbjct: 73 GFCYEDSLQPNKVKGKLVYCKLGNWGTEGV---VKKFGGIGSIMESD--QYPDLAQIFMA 237
Query: 442 PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYV 501
P+ I+N GE V YIKS S ++ I +K P V +SSRGP+ V
Sbjct: 238 PATILNHTIGESVTNYIKSTRSPSAVIYKTHEEKCP-----APFVATFSSRGPNPGSHNV 402
Query: 502 LKPDITAPGTSILAAWPTNVPVSNF-GTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNG 560
LKPDI APG ILA++ ++ G F+ F+L+ GTSM+CPHVAGVAA +K H
Sbjct: 403 LKPDIAAPGIDILASYTLRKSITGSEGDTQFSEFSLLSGTSMACPHVAGVAAYVKSFHPN 582
Query: 561 WSPSSIRSAIMTTS 574
W+P++IRSAI+TT+
Sbjct: 583 WTPAAIRSAIITTA 624
>TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,
partial (10%)
Length = 574
Score = 99.0 bits (245), Expect = 3e-21
Identities = 53/135 (39%), Positives = 79/135 (58%), Gaps = 4/135 (2%)
Frame = +2
Query: 273 DIIAAIDAAISDGVDILSISLGSDDLL----LYKDPVAIATFAAMEKGIFVSTSAGNNGP 328
D++AAID AI+DGVD+ S+S G ++ ++ D V+I + A+ K I V SAGN+GP
Sbjct: 2 DVLAAIDQAITDGVDVNSVSAGGRSIISAEEIFTDEVSIGSVHALAKNILVVASAGNDGP 181
Query: 329 SFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFSANNFPIVFMGMCD 388
+ ++ N PWV TVAA T+DREF T+TLGN +TG S ++ F ++ +
Sbjct: 182 ALGTVLNVAPWVFTVAASTIDREFSSTITLGNNQQITGASLFVNLPPNQLFSLI---LST 352
Query: 389 NVKELNTVKRKIVVC 403
+ K N R +C
Sbjct: 353 DAKLANATNRDAQLC 397
>TC19209 similar to GB|BAB11244.1|10177874|AB010074 serine protease-like
protein {Arabidopsis thaliana;} , partial (12%)
Length = 553
Score = 96.7 bits (239), Expect = 1e-20
Identities = 53/147 (36%), Positives = 78/147 (53%), Gaps = 7/147 (4%)
Frame = +1
Query: 25 LSDNYIIHMNLSDMPK-SFSNQHSWYESTLAQVTTTNNNLNNSTSS----KIFYTYTNVM 79
L YII ++ S MP +FS WY S + V + + +S +I Y+Y
Sbjct: 109 LKKTYIIQIDNSAMPAGTFSTHLEWYSSKVKSVLSKSLESETISSEEEEERIIYSYHTAF 288
Query: 80 NGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGA--WPTSDFGKD 137
+G +A L+PEE + L++ G ++ P+ +L TT SP FLGL + W D
Sbjct: 289 HGVAAKLTPEEAQKLESQEGVVAIFPETRYQLHTTRSPTFLGLEKMQSTEIWSEKLSNHD 468
Query: 138 IIVGVIDTGVWPESESFRDDGMTKIPS 164
++VGV DTG+WPES+SF D G +PS
Sbjct: 469 VVVGVSDTGIWPESQSFNDTGFKPVPS 549
>AV418734
Length = 287
Score = 94.7 bits (234), Expect = 5e-20
Identities = 50/103 (48%), Positives = 63/103 (60%)
Frame = +1
Query: 506 ITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSS 565
+TAPG +ILAAW P + N FN++ GTSM+CPHV G+A L+K H WSPS+
Sbjct: 4 VTAPGLNILAAWS---PAAG------NMFNIVSGTSMACPHVTGIATLVKAVHPSWSPSA 156
Query: 566 IRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNR 608
I+SAIMTT+ ILD HI R A F G+G +NP R
Sbjct: 157 IKSAIMTTATILDKHHRHI-SADPEQRTANAFDYGSGFVNPAR 282
>TC15026 weakly similar to UP|P93205 (P93205) Serine protease, SBT2 ,
partial (18%)
Length = 635
Score = 89.0 bits (219), Expect = 3e-18
Identities = 52/144 (36%), Positives = 76/144 (52%), Gaps = 3/144 (2%)
Frame = +1
Query: 616 YDIGVQDYINLLCALNFTQKNISAITRSSFNDCSKPSL---DLNYPSFIAFSNARNSSRT 672
YDI Q+Y + LC T + + S C DLNYP+ I+ + +S +
Sbjct: 1 YDIAPQEYFDFLCIQKLTPSQVQVFAKHSNRTCKHTLASAGDLNYPA-ISVVFPQKASIS 177
Query: 673 TNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQK 732
HRT TNVG + Y +++P KG V V P+ L F +K +K++YK+ T++
Sbjct: 178 VLTVHRTATNVGPAVSKYHVNVSPFKGASVKVEPDTLNFTRKYQKLAYKVTF--TVKTRQ 351
Query: 733 NKVAFGYLSWRDGKHVVRSPIVVT 756
+ FG L W+DG H VRSPIV+T
Sbjct: 352 IEPEFGGLVWKDGVHKVRSPIVIT 423
>TC13364 similar to UP|Q9LUM3 (Q9LUM3) Subtilisin proteinase-like protein,
partial (13%)
Length = 514
Score = 87.4 bits (215), Expect = 8e-18
Identities = 47/120 (39%), Positives = 67/120 (55%), Gaps = 2/120 (1%)
Frame = +1
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSP 88
+I+ + P F WYES+L+ ++TT + N +I YTY V +GFS LSP
Sbjct: 169 FIVQVQHEAKPSIFPTHKHWYESSLSSISTTTTSTN-----QIIYTYDTVFHGFSVKLSP 333
Query: 89 EEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLN--PYRGAWPTSDFGKDIIVGVIDTG 146
E + L++ S + IP+ +L TT SP FLGL G +DFG D+++GVIDTG
Sbjct: 334 LEAQKLQSLSHVTTLIPEQVRQLHTTRSPHFLGLKTADRAGLLHETDFGSDLVIGVIDTG 513
>AV426475
Length = 300
Score = 85.1 bits (209), Expect = 4e-17
Identities = 46/98 (46%), Positives = 61/98 (61%), Gaps = 5/98 (5%)
Frame = +2
Query: 463 SNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW----- 517
S+ + A + F T VK +P V +SSRGP+ P +LKPD+ APG +ILA W
Sbjct: 17 SSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIG 196
Query: 518 PTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLK 555
PT +PV +FN+I GTSMSCPHV+G+AA+LK
Sbjct: 197 PTGLPVDTRHV----SFNIISGTSMSCPHVSGLAAILK 298
>BP042306
Length = 549
Score = 81.6 bits (200), Expect = 4e-16
Identities = 43/100 (43%), Positives = 57/100 (57%), Gaps = 2/100 (2%)
Frame = +3
Query: 71 IFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGL--NPYRGA 128
I Y+Y +NGF+A L EE + +S KL TT S +FLGL N A
Sbjct: 240 ILYSYNKHINGFAALLEEEEAARIAKNPKVVSVFVSKEHKLHTTRSWEFLGLRGNGGNSA 419
Query: 129 WPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKG 168
W FG++ I+ IDTGVWPES+SF D G+ +P+KW+G
Sbjct: 420 WQKGRFGENTIIANIDTGVWPESQSFSDKGIGSVPAKWRG 539
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,623,062
Number of Sequences: 28460
Number of extensions: 169627
Number of successful extensions: 1012
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 940
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 949
length of query: 762
length of database: 4,897,600
effective HSP length: 97
effective length of query: 665
effective length of database: 2,136,980
effective search space: 1421091700
effective search space used: 1421091700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144727.4


