

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144726.1 + phase: 0
(240 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
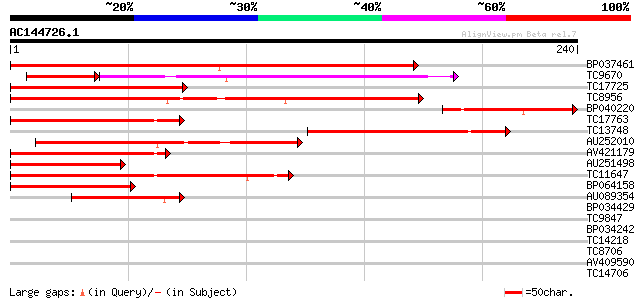
Score E
Sequences producing significant alignments: (bits) Value
BP037461 187 2e-48
TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%) 100 7e-31
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 116 4e-27
TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, pa... 113 3e-26
BP040220 101 1e-22
TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor,... 90 3e-19
TC13748 similar to UP|Q39401 (Q39401) MADS5 protein, partial (28%) 90 4e-19
AU252010 87 3e-18
AV421179 84 2e-17
AU251498 82 1e-16
TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, pa... 75 1e-14
BP064158 70 4e-13
AU089354 45 1e-05
BP034429 37 0.003
TC9847 similar to UP|AAR14272 (AAR14272) Predicted protein, part... 32 0.074
BP034242 28 1.1
TC14218 similar to UP|Q42896 (Q42896) Fructokinase , partial (77%) 28 1.8
TC8706 weakly similar to UP|Q7X9D9 (Q7X9D9) ABRH1 (Fragment), pa... 27 3.1
AV409590 27 4.1
TC14706 weakly similar to UP|AAQ72568 (AAQ72568) Ripening-relate... 27 4.1
>BP037461
Length = 551
Score = 187 bits (474), Expect = 2e-48
Identities = 95/174 (54%), Positives = 135/174 (76%), Gaps = 1/174 (0%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 8 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 187
Query: 61 SCMEKILERYERYSYAERQLVANDSES-QGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
S M K LERY++ +Y + + E+ + + EY +LKA+ + LQR+ R+ MGEDLG
Sbjct: 188 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 367
Query: 120 MSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIK 173
++ KEL+SLE+QLD++LK IR+ R Q M + +S+LQ+KE ++ E N L ++++
Sbjct: 368 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE 529
>TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%)
Length = 1060
Score = 100 bits (248), Expect(2) = 7e-31
Identities = 60/155 (38%), Positives = 88/155 (56%), Gaps = 3/155 (1%)
Frame = +3
Query: 39 CDAEVALIVFSHKGKLFEYATDSCMEKILERYERYSYAERQLVANDSESQGN---WTIEY 95
CDAEVALIVFS +G+L+EYA +S I +RY A S S+ N + E
Sbjct: 93 CDAEVALIVFSSRGRLYEYANNSVKATI----DRYKKACSDSSGAGSASEANAQFYQQEA 260
Query: 96 TRLKAKIDLLQRNYRHYMGEDLGSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQ 155
+L+ +I LQ N R MGE LGSM+ KEL++LE +L+ + IR+++N++++ I +Q
Sbjct: 261 DKLRVQISNLQNNNRQMMGESLGSMNAKELKNLETKLEKGISRIRSKKNELLFAEIEYMQ 440
Query: 156 KKEKVIQEQNNMLAKKIKEKEKIAAQQQAQWEHPN 190
K+E + N +L KI E E+ HPN
Sbjct: 441 KREIDLHNNNQLLRAKIAESER---------NHPN 518
Score = 49.7 bits (117), Expect(2) = 7e-31
Identities = 24/31 (77%), Positives = 27/31 (86%)
Frame = +1
Query: 8 LKRIENKINRQVTFSKRRAGLLKKAHEISVL 38
+KRIEN NRQVTF KRR GLLKKA+E+SVL
Sbjct: 1 IKRIENTTNRQVTFCKRRNGLLKKAYELSVL 93
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 116 bits (290), Expect = 4e-27
Identities = 57/75 (76%), Positives = 67/75 (89%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALIVFS +GKL+E+ +
Sbjct: 276 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSTRGKLYEFCSS 455
Query: 61 SCMEKILERYERYSY 75
S M K LERY++ SY
Sbjct: 456 SSMVKTLERYQKCSY 500
>TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, partial
(58%)
Length = 1100
Score = 113 bits (283), Expect = 3e-26
Identities = 66/178 (37%), Positives = 109/178 (61%), Gaps = 3/178 (1%)
Frame = +2
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++K IEN NRQVT+SKRR G+ KKAHE+SVLCDA+V+LI+FS K+ EY +
Sbjct: 137 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYISP 316
Query: 61 SCMEK-ILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGE--DL 117
K I+++Y++ + + L + E +LK + L+R RH +GE D+
Sbjct: 317 GLTTKRIIDQYQK-TLGDIDLWRSHYEKM---LENLKKLKEINNKLRRQIRHRLGEGLDM 484
Query: 118 GSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEK 175
+S ++L+ LE+ + +++ IR R+ V+ +KK + +++ N L ++KEK
Sbjct: 485 DDLSFQQLRKLEEDMVSSIGKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELKEK 658
>BP040220
Length = 541
Score = 101 bits (251), Expect = 1e-22
Identities = 46/58 (79%), Positives = 53/58 (91%), Gaps = 1/58 (1%)
Frame = -2
Query: 184 AQWEHPNHHGVNPNYLLQQQLPTLNMGGNYREE-APEMGRNELDLTLEPLYTCHLGCF 240
AQW+HPNH GVN +L+QQ LPTLNMGGNYRE+ AP++GRNELDLTLEPLY+CHLGCF
Sbjct: 540 AQWDHPNH-GVNAPFLMQQPLPTLNMGGNYREDHAPDVGRNELDLTLEPLYSCHLGCF 370
>TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor, partial
(35%)
Length = 737
Score = 90.1 bits (222), Expect = 3e-19
Identities = 46/74 (62%), Positives = 59/74 (79%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M R ++Q+K+I+N RQVTFSKRR GL KKA E+SVLCDA+VAL+VFS GKLFEY+
Sbjct: 501 MAREKIQIKKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALVVFSSTGKLFEYSNL 680
Query: 61 SCMEKILERYERYS 74
S M++ILER+ +S
Sbjct: 681 S-MKEILERHHLHS 719
>TC13748 similar to UP|Q39401 (Q39401) MADS5 protein, partial (28%)
Length = 624
Score = 89.7 bits (221), Expect = 4e-19
Identities = 48/86 (55%), Positives = 64/86 (73%)
Frame = +1
Query: 127 SLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAAQQQAQW 186
+LEQQLD+ALK IR+R+NQ ++ESISELQKK+K +QEQNN+L KKIKEKEK Q+ Q
Sbjct: 1 NLEQQLDSALKHIRSRKNQALFESISELQKKDKALQEQNNLLTKKIKEKEKAITTQEEQE 180
Query: 187 EHPNHHGVNPNYLLQQQLPTLNMGGN 212
N V + L+ Q L ++N+GG+
Sbjct: 181 GLQNSANV-ASGLVTQPLESMNIGGS 255
>AU252010
Length = 328
Score = 86.7 bits (213), Expect = 3e-18
Identities = 51/114 (44%), Positives = 72/114 (62%), Gaps = 1/114 (0%)
Frame = +1
Query: 12 ENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATDS-CMEKILERY 70
EN NRQVT+SKR+ G+LKKA EI+VLCDA+V+LI+F+ GK+ +Y + S + +LERY
Sbjct: 1 ENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISPSTTLVDMLERY 180
Query: 71 ERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSMSLKE 124
+ S +R A G E RLK + D +Q RH G+D+ S++ KE
Sbjct: 181 HKTS-GKRLWDAKHENLNG----EIERLKKENDGMQIELRHLKGDDINSLNYKE 327
>AV421179
Length = 207
Score = 84.3 bits (207), Expect = 2e-17
Identities = 42/68 (61%), Positives = 55/68 (80%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M R R+Q+K+I+N +RQVTFSKRR GL KKA E+S LCDA++ALIVFS KLFEYA+
Sbjct: 6 MTRKRIQIKKIDNISSRQVTFSKRRKGLFKKAQELSTLCDADIALIVFSATNKLFEYASS 185
Query: 61 SCMEKILE 68
S ++K++E
Sbjct: 186S-IQKVIE 206
>AU251498
Length = 342
Score = 81.6 bits (200), Expect = 1e-16
Identities = 39/49 (79%), Positives = 45/49 (91%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFS 49
MGRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVALIVFS
Sbjct: 45 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS 191
>TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, partial (25%)
Length = 843
Score = 74.7 bits (182), Expect = 1e-14
Identities = 43/121 (35%), Positives = 75/121 (61%), Gaps = 1/121 (0%)
Frame = +1
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
+GR ++++K++ N+ N QVTFSKRR+GL KKA E+ +LCD E+AL+VFS K+F +
Sbjct: 85 LGRQKIEMKKMTNESNLQVTFSKRRSGLFKKASELCILCDVEIALVVFSPGEKVFSFGHP 264
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLK-AKIDLLQRNYRHYMGEDLGS 119
S +E +++RY + + E+Q N + ++ ++I+ L R++ GE+L
Sbjct: 265 S-VEAVIKRYFSQAPPQTSDTMQYIEAQRNSKVHELNVELSQINNLLDTVRNH-GEELNR 438
Query: 120 M 120
+
Sbjct: 439 L 441
>BP064158
Length = 394
Score = 69.7 bits (169), Expect = 4e-13
Identities = 33/53 (62%), Positives = 44/53 (82%)
Frame = +3
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGK 53
MGR ++Q +++EN NRQVTFSKRR GL+KKA+E+SVLC +VALI+FS G+
Sbjct: 165 MGRVKLQNQKVENTTNRQVTFSKRRNGLIKKAYELSVLCXFDVALIMFSPSGR 323
>AU089354
Length = 178
Score = 45.1 bits (105), Expect = 1e-05
Identities = 24/49 (48%), Positives = 34/49 (68%), Gaps = 1/49 (2%)
Frame = +3
Query: 27 GLLKKAHEISVLCDAEVALIVFSHKGKLFEYATDSCME-KILERYERYS 74
G+LKKA EI+VLC A+V+LI F+ GK+ +Y S ++LERY + S
Sbjct: 3 GILKKAIEITVLCXAQVSLIXFAASGKMHDYIRPSITXVEMLERYXKTS 149
>BP034429
Length = 567
Score = 37.0 bits (84), Expect = 0.003
Identities = 18/63 (28%), Positives = 35/63 (54%)
Frame = -1
Query: 110 RHYMGEDLGSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLA 169
R GED + +L LE+ L + LK + + + + + I+ +QKK ++E+N +L
Sbjct: 504 RRMTGEDFEGLEFDDLLELEKTLPSGLKRVIELKEKRIMDEITAVQKKGIQLEEENKLLK 325
Query: 170 KKI 172
+K+
Sbjct: 324 QKM 316
>TC9847 similar to UP|AAR14272 (AAR14272) Predicted protein, partial (73%)
Length = 952
Score = 32.3 bits (72), Expect = 0.074
Identities = 24/99 (24%), Positives = 48/99 (48%), Gaps = 2/99 (2%)
Frame = +1
Query: 91 WTIEYTRLKAKIDLLQRNYRHYMGEDLGSMSLKELQSLEQQLDTALKLIRTRRNQVMYES 150
W I R +D+++ R + E + +E+ L +QLD + + + Q++ E
Sbjct: 463 WLIGKKRKLVNVDVVESMRRIAIQEM--NRKDREIDGLNEQLDEDSRCLEHLQIQLVEER 636
Query: 151 I--SELQKKEKVIQEQNNMLAKKIKEKEKIAAQQQAQWE 187
+ +++ ++QEQ NML ++E E + + Q Q E
Sbjct: 637 SKRARVERDNAMLQEQVNMLMNMLQETEPMGDEDQDQEE 753
>BP034242
Length = 539
Score = 28.5 bits (62), Expect = 1.1
Identities = 27/112 (24%), Positives = 50/112 (44%), Gaps = 8/112 (7%)
Frame = -1
Query: 42 EVALIVFSHKGKLFEYATDSCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAK 101
E ++F+ G L+++ TD +I+ R R A+R + A E+ Y LK +
Sbjct: 509 EDLFLIFASDG-LWDHLTDEAAVEIISRSPRTGIAKRLVRAALEEAAKKNGKRYEDLK-R 336
Query: 102 IDLLQRNYRH--------YMGEDLGSMSLKELQSLEQQLDTALKLIRTRRNQ 145
I+ R H Y+ LGS++ Q + + +DT + + R ++
Sbjct: 335 IEKGVRRLFHDDITVIVLYLDNSLGSLNSSRHQGVYRCIDTPVDIFSLRADE 180
>TC14218 similar to UP|Q42896 (Q42896) Fructokinase , partial (77%)
Length = 1170
Score = 27.7 bits (60), Expect = 1.8
Identities = 15/53 (28%), Positives = 31/53 (58%)
Frame = -3
Query: 126 QSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKI 178
QSL +L + L +++ ++ + + K K IQE N++L +++K K+K+
Sbjct: 1093 QSLISKLGS*HYLAKSQST*SCRQNTEDQEMKTKEIQETNSLLNQQLKRKKKV 935
>TC8706 weakly similar to UP|Q7X9D9 (Q7X9D9) ABRH1 (Fragment), partial
(49%)
Length = 1084
Score = 26.9 bits (58), Expect = 3.1
Identities = 18/64 (28%), Positives = 34/64 (53%), Gaps = 5/64 (7%)
Frame = +1
Query: 124 ELQSLEQQLDTALKLIRTRRNQVMYESIS----ELQKKEKVIQEQNNMLAK-KIKEKEKI 178
EL+S+ + K+ ++R + E+ L K EK+ +E NN LAK ++ E +++
Sbjct: 508 ELESIVKIKQAEAKMFQSRSDDARREAEGLKRISLAKSEKIEEEYNNRLAKLRLVETDEM 687
Query: 179 AAQQ 182
Q+
Sbjct: 688 RKQK 699
>AV409590
Length = 396
Score = 26.6 bits (57), Expect = 4.1
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +3
Query: 170 KKIKEKEKIAAQQQAQWEHP 189
K++KEK+K+AAQQ A+ P
Sbjct: 318 KRLKEKQKLAAQQAAEKGKP 377
>TC14706 weakly similar to UP|AAQ72568 (AAQ72568) Ripening-related protein,
partial (39%)
Length = 701
Score = 26.6 bits (57), Expect = 4.1
Identities = 9/38 (23%), Positives = 20/38 (51%)
Frame = +3
Query: 66 ILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKID 103
+ Y++++ + + ND WT+EY +L+ I+
Sbjct: 303 VSSHYKKFTLIFQVIENNDGHDAVKWTVEYEKLREDIE 416
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,427,529
Number of Sequences: 28460
Number of extensions: 37503
Number of successful extensions: 283
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 276
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 278
length of query: 240
length of database: 4,897,600
effective HSP length: 88
effective length of query: 152
effective length of database: 2,393,120
effective search space: 363754240
effective search space used: 363754240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC144726.1


