

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.15 + phase: 0
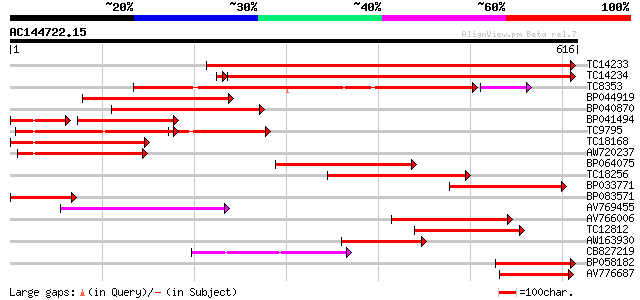
(616 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%) 455 e-128
TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%) 427 e-121
TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa pro... 322 1e-91
BP044919 271 2e-73
BP040870 260 4e-70
BP041494 171 4e-64
TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment),... 221 3e-58
TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat... 211 2e-55
AW720237 192 2e-49
BP064075 171 4e-43
TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, p... 148 3e-36
BP033771 139 1e-33
BP083571 127 4e-30
AV769455 122 2e-28
AV766006 119 1e-27
TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein ... 107 4e-24
AW163930 100 7e-22
CB827219 99 3e-21
BP058182 92 3e-19
AV776687 91 5e-19
>TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%)
Length = 1570
Score = 455 bits (1170), Expect = e-128
Identities = 235/403 (58%), Positives = 307/403 (75%), Gaps = 3/403 (0%)
Frame = +1
Query: 215 VSLLTLKGDSFDVKATSGDTHLGGEDFDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTA 274
VSLLT++ F+VKAT+GDTHLGGEDFDNRMV+H V+EF+RKNK D+SGN RALRRL+TA
Sbjct: 1 VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTA 180
Query: 275 CERAKRTLSYDTEATIGIDAIYEGVDFHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKM 334
CERAKRTLS + TI ID++YEG+DF+ +TRA+FE++N DLF KC+E V+ CL +AKM
Sbjct: 181 CERAKRTLSSTAQTTIEIDSLYEGIDFYSPITRARFEEMNMDLFRKCMEPVEKCLRDAKM 360
Query: 335 DKNIVDDVVLVGGSSRIPKVQQLLQDFFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIK 393
DK V DVVLVGGS+RIPKVQQLLQDFF GK+L SINPDEAVAYGAAVQAA+L +G +
Sbjct: 361 DKRSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNE 540
Query: 394 NVPNLVLQDVTPLSLGTSIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGE 453
V +L+L DVTPLSLG +M V+IP+NT+IP +K+Q + T D+Q V I V+EGE
Sbjct: 541 KVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGE 720
Query: 454 RMVAIENNLLGLFNLK-VRRAPRGLP-IQVCFSIDADGILNVSAEEESSGNKKNITITNE 511
R +NNLLG F L + APRG+P I VCF IDA+GILNVSAE++++G K ITITN+
Sbjct: 721 RTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITND 900
Query: 512 NGRLTSEEIERMIQEAEHFKAEDMKHVKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVD 571
GRL+ ++IE+M+QEAE +K+ED +H KKV A NAL++Y Y+MR +KD+ +A KL S D
Sbjct: 901 KGRLSKDDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTVKDEKIAGKLDSAD 1080
Query: 572 KVRINSAMIKGKKLIDDNQPKETYVFVDFLRELKNTFDSALNK 614
K +I A+ + + ++ NQ E F D ++EL++ + + K
Sbjct: 1081KTKIEDAIEQAIQWLESNQLAEADEFEDKMKELESICNPIIAK 1209
>TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%)
Length = 1528
Score = 427 bits (1097), Expect(2) = e-121
Identities = 222/381 (58%), Positives = 286/381 (74%), Gaps = 3/381 (0%)
Frame = +3
Query: 237 GGEDFDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIY 296
GGEDFDNRMV+H V+EF+RKNK D+SGN RALRRL+TACERAKRTLS + TI ID++Y
Sbjct: 48 GGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLY 227
Query: 297 EGVDFHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQ 356
EG+DF+ +TRA+FE+LN DLF KC+E V+ CL +AKMDK V DVVLVGGS+RIPKVQQ
Sbjct: 228 EGIDFYSPITRARFEELNMDLFRKCMEPVEKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQ 407
Query: 357 LLQDFFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDD 415
LLQDFF GK+L SINPDEAVAYGAAVQAA+L +G + V +L+L DVTPLSLG
Sbjct: 408 LLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGG 587
Query: 416 IMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAP 474
+M V+IP+NT+IP +K+Q + T D+Q V I VYEGER +NNLLG F L + AP
Sbjct: 588 VMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAP 767
Query: 475 RGLP-IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAE 533
RG+P I VCF IDA+GILNVSAE++++G K ITITN+ GRL+ E+IE+M+QEAE +K+E
Sbjct: 768 RGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSE 947
Query: 534 DMKHVKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPKE 593
D +H KKV A NAL++Y Y+MR +KD+ + KL DK +I A+ + + +D NQ E
Sbjct: 948 DEEHKKKVEAKNALENYAYNMRNTIKDEKIGGKLDPADKKKIEDAIEQAIQWLDSNQLAE 1127
Query: 594 TYVFVDFLRELKNTFDSALNK 614
F D ++EL++ + + K
Sbjct: 1128ADEFEDKMKELESICNPIIAK 1190
Score = 26.2 bits (56), Expect(2) = e-121
Identities = 10/12 (83%), Positives = 12/12 (99%)
Frame = +1
Query: 225 FDVKATSGDTHL 236
F+VKAT+GDTHL
Sbjct: 13 FEVKATAGDTHL 48
>TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (74%)
Length = 1752
Score = 322 bits (824), Expect(2) = 1e-91
Identities = 185/380 (48%), Positives = 248/380 (64%), Gaps = 6/380 (1%)
Frame = +3
Query: 135 EIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAAGLAYGLQKR 194
E AE +L V AV+TVPAYFND+ R+ATKDAG IAGL+V RIINEPTAA L+YG+ +
Sbjct: 3 ETAEAYLGKSVSKAVITVPAYFNDAHRQATKDAGRIAGLDVQRIINEPTAAALSYGMTNK 182
Query: 195 ESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRMVHHLVKEFR 254
E I +FDLGGGTFDVS+L + F+VKAT+GDT LGGEDFDN ++ LV EF+
Sbjct: 183 EGLIA-----VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVDEFK 347
Query: 255 RKNKVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVD----FHLLVTRAKF 310
R +DLS + AL+RL+ A E+AK LS ++ I + I ++ +TR+KF
Sbjct: 348 RTESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKF 527
Query: 311 EQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMGKDLYNS 370
E L L E+ KNCL +A + VD+V+LVGG +R+PKVQ+++ F GK
Sbjct: 528 EALVNHLIERTKAPCKNCLKDANISIKEVDEVLLVGGMTRVPKVQEVVSAIF-GKSPSKG 704
Query: 371 INPDEAVAYGAAVQAALLCDGIKNVPNLVLQDVTPLSLGTSIYDDIMDVVIPKNTSIPIR 430
+NPDEAVA GAA+Q +L +K L+L DVTPLSLG I +I +NT+IP +
Sbjct: 705 VNPDEAVAMGAAIQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTK 875
Query: 431 KKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNL-KVRRAPRGLP-IQVCFSIDAD 488
K Q + T D+Q+ V I V +GER +A +N LG F L + APRGLP I+V F IDA+
Sbjct: 876 KSQVFSTAADNQTQVGIKVLQGEREMASDNKSLGEFELVGIPPAPRGLPQIEVTFDIDAN 1055
Query: 489 GILNVSAEEESSGNKKNITI 508
GI+ VSA+++S+G ++ ITI
Sbjct: 1056GIVTVSAKDKSTGKEQQITI 1115
Score = 32.3 bits (72), Expect(2) = 1e-91
Identities = 16/58 (27%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Frame = +2
Query: 512 NGRLTSEEIERMIQEAEHFKAEDMKHVKKVRAMNALDDYLYDMRKVMKD--DSVASKL 567
+G L+ +EIE+M++EAE +D + + N+ D +Y + K + + D + S++
Sbjct: 1124 SGGLSDDEIEKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEV 1297
>BP044919
Length = 519
Score = 271 bits (693), Expect = 2e-73
Identities = 133/164 (81%), Positives = 151/164 (91%)
Frame = +3
Query: 80 GRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEISSMVLTKMREIAEK 139
GRK+SDSVIQ D++LWPFKV++G DDKP I+VK+KGEEK+L EEISSMVL+KM+EIAE
Sbjct: 27 GRKYSDSVIQEDVKLWPFKVIAGTDDKPMIVVKHKGEEKKLVPEEISSMVLSKMKEIAEA 206
Query: 140 FLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPTAAGLAYGLQKRESNIG 199
FLES VKNAVVTVPAYFND+QRKATKDAG IAGLNVMRIINEPTAA LAYGLQKR + +G
Sbjct: 207 FLESSVKNAVVTVPAYFNDAQRKATKDAGTIAGLNVMRIINEPTAAALAYGLQKRANLVG 386
Query: 200 KRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDN 243
+RNVFIFDLGGGTFDVSLLT+K D+F+VKATSGDTHLG +DFDN
Sbjct: 387 ERNVFIFDLGGGTFDVSLLTIKDDNFEVKATSGDTHLGXKDFDN 518
>BP040870
Length = 505
Score = 260 bits (665), Expect = 4e-70
Identities = 127/167 (76%), Positives = 150/167 (89%)
Frame = +2
Query: 111 VKYKGEEKRLCAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVI 170
V YKGEEK+ AEEISSMVL KM+EIAE +L + +KNAVVTVPAYFNDSQR+ATKDAGVI
Sbjct: 5 VNYKGEEKQFSAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVI 184
Query: 171 AGLNVMRIINEPTAAGLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKAT 230
+GLNVMRIINEPTAA +AYGL K+ ++ G++NV IFDLGGGTFDVSLLT++ F+VK+T
Sbjct: 185 SGLNVMRIINEPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKST 364
Query: 231 SGDTHLGGEDFDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACER 277
+GDTHLGGEDFDNRMV+H V+EF+RKNK D+SGNARALRRL+TACER
Sbjct: 365 AGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACER 505
>BP041494
Length = 572
Score = 171 bits (434), Expect(2) = 4e-64
Identities = 86/110 (78%), Positives = 96/110 (87%)
Frame = +3
Query: 74 DAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEISSMVLTKM 133
DAKRLIGRKFSD +++DM+LWPFKV+SG D+P I+V YK EEK+ AEEISSMVL KM
Sbjct: 243 DAKRLIGRKFSDDSVESDMKLWPFKVISGPVDRPMIVVNYKNEEKQFTAEEISSMVLIKM 422
Query: 134 REIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPT 183
REIAE +L S VKN VVTVPAYFNDSQR+ATKDAGVIAGLNVMRIINEPT
Sbjct: 423 REIAEAYLGSTVKNVVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 572
Score = 90.9 bits (224), Expect(2) = 4e-64
Identities = 46/66 (69%), Positives = 52/66 (78%)
Frame = +1
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
M + + PAIGIDLGTTYSCV VWQ N+ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 61 MVGEGESPAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDFERLIGDAA 234
Query: 61 KNQAAS 66
KNQ S
Sbjct: 235 KNQMRS 252
>TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment), partial
(63%)
Length = 981
Score = 221 bits (563), Expect = 3e-58
Identities = 118/178 (66%), Positives = 138/178 (77%), Gaps = 1/178 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNRITPS+VAF + +RLIG+AAKNQ A
Sbjct: 145 GTVIGIDLGTTYSCVGVYR--NGHVEIIANDQGNRITPSWVAFNDGERLIGEAAKNQFAV 318
Query: 67 NPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYK-GEEKRLCAEEI 125
NP T+FD KRLIGRKF D +Q DM+L P+K+V+ D KP I VK K GE K EE+
Sbjct: 319 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQVKIKDGETKVFSPEEV 495
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNVMRIINEPT 183
S+M+LTKM+E AE FL + +AVVTVPAYFND+QR+ATKDAGVIAGLNV RIINEPT
Sbjct: 496 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 669
Score = 113 bits (282), Expect = 1e-25
Identities = 55/111 (49%), Positives = 79/111 (70%)
Frame = +2
Query: 173 LNVMRIINEPTAAGLAYGLQKRESNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSG 232
L ++ + P AA +AYGL K+ G++N+ +FDLGGGTFDVS+LT+ F+V AT+G
Sbjct: 638 LMLLELSMNPPAAAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNG 808
Query: 233 DTHLGGEDFDNRMVHHLVKEFRRKNKVDLSGNARALRRLKTACERAKRTLS 283
DTHLGGEDFD R++ + +K ++K+ D+S + RAL +L+ ERAKR LS
Sbjct: 809 DTHLGGEDFDQRIMEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALS 961
>TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (23%)
Length = 508
Score = 211 bits (538), Expect = 2e-55
Identities = 109/152 (71%), Positives = 125/152 (81%)
Frame = +2
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++ EII NDQGNR TPS+VAFT+ +RLIGDAA
Sbjct: 59 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 232
Query: 61 KNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRL 120
KNQ A NP NTVFDAKRLIGR+ SD+ +Q+DM+LWPFKV SG +KP I V YKGE+K+
Sbjct: 233 KNQVAMNPINTVFDAKRLIGRRVSDASVQSDMKLWPFKVNSGPAEKPMIQVSYKGEDKQF 412
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTV 152
AEEISSMVL KMREIAE +L S +KNAVVTV
Sbjct: 413 AAEEISSMVLMKMREIAEAYLGSTIKNAVVTV 508
>AW720237
Length = 459
Score = 192 bits (487), Expect = 2e-49
Identities = 95/141 (67%), Positives = 110/141 (77%)
Frame = +3
Query: 9 AIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAAKNQAASNP 68
AIGIDLGTTYSCV VW N+ EII NDQGNR TPS+V FT +RLIGDAAKNQ A NP
Sbjct: 42 AIGIDLGTTYSCVGVWM--NDRVEIIANDQGNRTTPSYVGFTESERLIGDAAKNQVARNP 215
Query: 69 ANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKGEEKRLCAEEISSM 128
NTVFDAKRLIGRKF D ++Q D++LWPFKV +G D KP I VKYKGE K+ AE++SSM
Sbjct: 216 INTVFDAKRLIGRKFDDPIVQKDIKLWPFKVEAGPDGKPLIAVKYKGETKKFHAEQVSSM 395
Query: 129 VLTKMREIAEKFLESPVKNAV 149
VLTKM+E AE +L +K+ V
Sbjct: 396 VLTKMKETAEAYLNKTIKDIV 458
>BP064075
Length = 554
Score = 171 bits (432), Expect = 4e-43
Identities = 84/155 (54%), Positives = 118/155 (75%), Gaps = 1/155 (0%)
Frame = -2
Query: 289 TIGIDAIYEGVDFHLLVTRAKFEQLNRDLFEKCLEIVKNCLANAKMDKNIVDDVVLVGGS 348
TI +D +Y+G+DF+ +TRAKFE+LN+D F+KC+E+V+ C+ ++KMDK+ + DVVLVGGS
Sbjct: 553 TIELDFLYQGIDFYSSITRAKFEELNKDYFQKCMELVEKCVIDSKMDKSNIHDVVLVGGS 374
Query: 349 SRIPKVQQLLQDFFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLS 407
SRIPKV+QLL DFF KDL N INPDEAVA+GAAV A++L + + V +L+L++VTPLS
Sbjct: 373 SRIPKVRQLLMDFFGRKDLCNRINPDEAVAHGAAVHASILSGEFSEKVQSLLLREVTPLS 194
Query: 408 LGTSIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQ 442
LG + IM+ +IP+NT IP + T + +Q
Sbjct: 193 LGLEKHGGIMETIIPRNTMIPTTMDHVFTTHFHNQ 89
>TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, partial
(24%)
Length = 476
Score = 148 bits (373), Expect = 3e-36
Identities = 79/158 (50%), Positives = 111/158 (70%), Gaps = 3/158 (1%)
Frame = +3
Query: 346 GGSSRIPKVQQLLQDFFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVT 404
GGS+RIPKVQQLL+D+F GK+ +NPDEAVAYGAAVQ ++L +G + +++L DV
Sbjct: 3 GGSTRIPKVQQLLKDYFEGKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVA 182
Query: 405 PLSLGTSIYDDIMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLG 464
PL+LG +M +IP+NT IP +K Q + T D Q++V+I V+EGER + + LLG
Sbjct: 183 PLTLGIETVGGVMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLG 362
Query: 465 LFNLK-VRRAPRGLP-IQVCFSIDADGILNVSAEEESS 500
F+L + APRG P I+V F +DA+GILNV AE++ +
Sbjct: 363 KFDLSGIPPAPRGTPQIEVTFEVDANGILNVKAEDKGT 476
>BP033771
Length = 513
Score = 139 bits (351), Expect = 1e-33
Identities = 69/128 (53%), Positives = 93/128 (71%)
Frame = -2
Query: 478 PIQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKH 537
P+ VCF +DA+GILNVSAEEE++G K IT+TN+NGRL+SE+I R+IQEAE +AED +
Sbjct: 512 PLNVCFDLDANGILNVSAEEETTGIKNGITVTNDNGRLSSEQIMRLIQEAEKHRAEDRMY 333
Query: 538 VKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPKETYVF 597
KKV AMNALDDY+Y +R +KD +SKL DK +I A+ K + L+ + ET VF
Sbjct: 332 EKKVEAMNALDDYVYKLRNAIKDIDRSSKLRPQDKSKIRVAVTKARNLLGTSDQTETEVF 153
Query: 598 VDFLRELK 605
D+L E++
Sbjct: 152 EDYLNEME 129
>BP083571
Length = 423
Score = 127 bits (320), Expect = 4e-30
Identities = 60/72 (83%), Positives = 67/72 (92%)
Frame = +1
Query: 1 MAKKHKGPAIGIDLGTTYSCVAVWQDQNNTAEIIHNDQGNRITPSFVAFTNDQRLIGDAA 60
MA K++GPAIGIDLGTTYSCVAVWQ+QN+ AEIIHN+QGNR TPSFVAFT+ QRLIGDAA
Sbjct: 145 MATKYEGPAIGIDLGTTYSCVAVWQEQNDRAEIIHNEQGNRTTPSFVAFTDTQRLIGDAA 324
Query: 61 KNQAASNPANTV 72
KNQAA+NP NTV
Sbjct: 325 KNQAATNPTNTV 360
>AV769455
Length = 556
Score = 122 bits (306), Expect = 2e-28
Identities = 68/184 (36%), Positives = 100/184 (53%), Gaps = 1/184 (0%)
Frame = +1
Query: 56 IGDAAKNQAASNPANTVFDAKRLIGRKFSDSVIQNDMELWPFKVVSGFDDKPEIIVKYKG 115
+G A + NP N++ KRLIGR+F+D +Q D++ PF V G D P I +Y G
Sbjct: 4 LGTAGASSTMMNPKNSISQLKRLIGRQFADPELQRDLKSLPFAVTEGPDGFPLIHARYLG 183
Query: 116 EEKRLCAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGVIAGLNV 175
E + ++ M+L+ ++EIA+K L + V + + +P YF D QR+A DA IAGL+
Sbjct: 184 ETRTFTPTQVFGMMLSNLKEIAQKNLNAAVVDCCIGIPIYFTDLQRRAVLDAATIAGLHP 363
Query: 176 MRIINEPTAAGLAYGLQKRE-SNIGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDT 234
+R+ +E TA LAYG+ K + NV D+G + V + K V A S D
Sbjct: 364 LRLFHETTATALAYGIYKTDLPENDPLNVAFVDVGHASMQVCIAGYKKGQLKVLAHSYDR 543
Query: 235 HLGG 238
LGG
Sbjct: 544 SLGG 555
>AV766006
Length = 408
Score = 119 bits (299), Expect = 1e-27
Identities = 65/133 (48%), Positives = 90/133 (66%), Gaps = 2/133 (1%)
Frame = -1
Query: 416 IMDVVIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAP 474
+M +I +NT IP RK Q + T D Q++V+I VYEGER + +N LG F+L + A
Sbjct: 399 VMSKIIERNTVIPTRKTQTFTTYQDKQTTVSIQVYEGERAMTKDNRQLGKFDLNGIPAAA 220
Query: 475 RGLP-IQVCFSIDADGILNVSAEEESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAE 533
RG P I+V F +DA+GIL VSAE+++S K+ ITITN+ GRL+ EEIERM++EAE F
Sbjct: 219 RGTPQIEVTFEVDANGILAVSAEDKASSAKEAITITNDKGRLSKEEIERMVKEAEEFADA 40
Query: 534 DMKHVKKVRAMNA 546
D + +KV N+
Sbjct: 39 DAEVKRKVEGKNS 1
>TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (48%)
Length = 746
Score = 107 bits (268), Expect = 4e-24
Identities = 62/122 (50%), Positives = 82/122 (66%), Gaps = 2/122 (1%)
Frame = +1
Query: 440 DDQSSVNIDVYEGERMVAIENNLLGLFNLK-VRRAPRG-LPIQVCFSIDADGILNVSAEE 497
D+Q+SV I VYEGE A +N LG F L AP+G I VCF +DADGI+ VSAE+
Sbjct: 1 DNQTSVWIKVYEGEGAKADKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED 180
Query: 498 ESSGNKKNITITNENGRLTSEEIERMIQEAEHFKAEDMKHVKKVRAMNALDDYLYDMRKV 557
S KK ITITN GRL+ EE+ RM+++A +KAED + KKV+A N+L++Y Y+ R
Sbjct: 181 MSLRLKKTITITNRLGRLSQEEMSRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDR 360
Query: 558 MK 559
+K
Sbjct: 361 VK 366
>AW163930
Length = 359
Score = 100 bits (249), Expect = 7e-22
Identities = 54/93 (58%), Positives = 68/93 (73%), Gaps = 1/93 (1%)
Frame = +1
Query: 361 FFMGKDLYNSINPDEAVAYGAAVQAALLC-DGIKNVPNLVLQDVTPLSLGTSIYDDIMDV 419
FF GK+L SINPDEAVAYGAAVQAA+L +G + V +L+L DVTPLSLG +M V
Sbjct: 40 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 219
Query: 420 VIPKNTSIPIRKKQEYVTCYDDQSSVNIDVYEG 452
+IP+NT+IP +K+Q + T D+Q V I VYEG
Sbjct: 220 LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEG 318
>CB827219
Length = 544
Score = 98.6 bits (244), Expect = 3e-21
Identities = 54/174 (31%), Positives = 97/174 (55%)
Frame = +3
Query: 198 IGKRNVFIFDLGGGTFDVSLLTLKGDSFDVKATSGDTHLGGEDFDNRMVHHLVKEFRRKN 257
+G R + F++G G DV++ G +KA +G T +GGED M+HHL+ +
Sbjct: 27 VGVRKLLYFNMGAGYCDVAVTVTAGGVSQIKALAGST-IGGEDLLQNMMHHLLPDCENIF 203
Query: 258 KVDLSGNARALRRLKTACERAKRTLSYDTEATIGIDAIYEGVDFHLLVTRAKFEQLNRDL 317
K +++ L+ A A LS + +D + +G+ + ++ +FE++N+++
Sbjct: 204 KNHGVKEIKSMGLLRVATLDAIHRLSSQKSVQVDVD-LGDGLKINKVIDCEEFEEVNKNV 380
Query: 318 FEKCLEIVKNCLANAKMDKNIVDDVVLVGGSSRIPKVQQLLQDFFMGKDLYNSI 371
FEKC ++ CL ++K++ V+DV++VGG S IP+V+ L+ + GK LY I
Sbjct: 381 FEKCESLIIQCLQDSKIEVEDVNDVIIVGGCSYIPRVKNLVTNICKGKKLYEGI 542
>BP058182
Length = 449
Score = 91.7 bits (226), Expect = 3e-19
Identities = 48/87 (55%), Positives = 61/87 (69%)
Frame = -3
Query: 528 EHFKAEDMKHVKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLID 587
E + +DMK KV AMN LD YLYDM K MKD V+SKL S+ + +INSA++KG L+D
Sbjct: 447 EDHEVDDMKFKNKVTAMNDLDLYLYDMNKSMKDKCVSSKLPSMVQDKINSAIVKGXSLLD 268
Query: 588 DNQPKETYVFVDFLRELKNTFDSALNK 614
D Q ET VFV+FL ELK+ F+ A+ K
Sbjct: 267 DGQQLETSVFVEFLMELKSIFEPAMAK 187
>AV776687
Length = 595
Score = 90.9 bits (224), Expect = 5e-19
Identities = 44/80 (55%), Positives = 60/80 (75%)
Frame = -1
Query: 533 EDMKHVKKVRAMNALDDYLYDMRKVMKDDSVASKLTSVDKVRINSAMIKGKKLIDDNQPK 592
+DM K +AMNALDDYLY+M K+MKD V+SKL +V+INSA++KGK L+D + +
Sbjct: 592 DDMMFKNKRKAMNALDDYLYNMNKLMKDTCVSSKLPPKVQVKINSAIVKGKNLLDGGKEQ 413
Query: 593 ETYVFVDFLRELKNTFDSAL 612
E +VFVD LRELK+ F+ A+
Sbjct: 412 EAFVFVDLLRELKSIFEPAM 353
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,850,563
Number of Sequences: 28460
Number of extensions: 85253
Number of successful extensions: 370
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 348
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 348
length of query: 616
length of database: 4,897,600
effective HSP length: 96
effective length of query: 520
effective length of database: 2,165,440
effective search space: 1126028800
effective search space used: 1126028800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144722.15


