

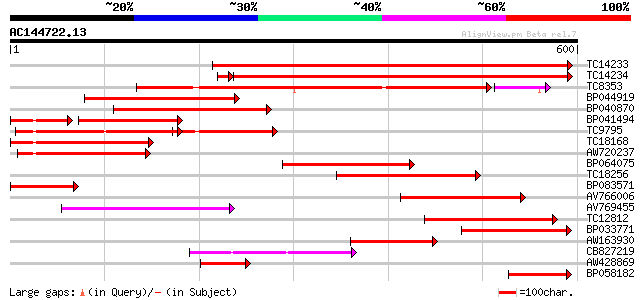
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.13 + phase: 0
(600 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%) 458 e-129
TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%) 427 e-122
TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa pro... 312 1e-89
BP044919 272 1e-73
BP040870 260 4e-70
BP041494 166 4e-64
TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment),... 220 4e-58
TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat... 215 1e-56
AW720237 197 5e-51
BP064075 173 6e-44
TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, p... 147 4e-36
BP083571 132 1e-31
AV766006 125 2e-29
AV769455 122 2e-28
TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein ... 112 2e-25
BP033771 111 3e-25
AW163930 101 3e-22
CB827219 86 2e-17
AW428869 73 1e-13
BP058182 73 2e-13
>TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%)
Length = 1570
Score = 458 bits (1178), Expect = e-129
Identities = 233/383 (60%), Positives = 295/383 (76%), Gaps = 2/383 (0%)
Frame = +1
Query: 215 VSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTA 274
VSLLTI+ +FEVKAT+GDTHLGGEDFDNRMVNHFV EF RK KDISG+PR LRRLRTA
Sbjct: 1 VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTA 180
Query: 275 CERTKRTLSFDTNATIDIDSLFEGIDFHSSITRAKFEQLNVDLFKKCIETVESCLSDAQI 334
CER KRTLS TI+IDSL+EGIDF+S ITRA+FE++N+DLF+KC+E VE CL DA++
Sbjct: 181 CERAKRTLSSTAQTTIEIDSLYEGIDFYSPITRARFEEMNMDLFRKCMEPVEKCLRDAKM 360
Query: 335 EKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQAALLC-EATE 393
+K SV D VLVGGS+RIPKVQQLL++FFN K++C SINPDEAVAYGAAVQAA+L E E
Sbjct: 361 DKRSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNE 540
Query: 394 KSLNLVLRDVTPLSLGTLVRGDLLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGE 453
K +L+L DVTPLSLG G +++V+IPRNT IP KK + + T D+ V ++V+EGE
Sbjct: 541 KVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGE 720
Query: 454 RLKASENNLLGKFRFS-IPPAPRGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNE 512
R + +NNLLGKF S IPPAPRG I VCF ID +GILNVSA + T G + +ITITN+
Sbjct: 721 RTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITND 900
Query: 513 KGRLSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIE 572
KGRLS ++IE+M+QEAE +K ED + KKKV+A N L++Y Y MR +KD+ ++ L S +
Sbjct: 901 KGRLSKDDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTVKDEKIAGKLDSAD 1080
Query: 573 RMKINSAMIKGMKLIDEKQHHEA 595
+ KI A+ + ++ ++ Q EA
Sbjct: 1081KTKIEDAIEQAIQWLESNQLAEA 1149
>TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%)
Length = 1528
Score = 427 bits (1099), Expect(2) = e-122
Identities = 219/361 (60%), Positives = 273/361 (74%), Gaps = 2/361 (0%)
Frame = +3
Query: 237 GGEDFDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLF 296
GGEDFDNRMVNHFV EF RK KDISG+PR LRRLRTACER KRTLS TI+IDSL+
Sbjct: 48 GGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLY 227
Query: 297 EGIDFHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQ 356
EGIDF+S ITRA+FE+LN+DLF+KC+E VE CL DA+++K SV D VLVGGS+RIPKVQQ
Sbjct: 228 EGIDFYSPITRARFEELNMDLFRKCMEPVEKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQ 407
Query: 357 LLKEFFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGD 415
LL++FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G
Sbjct: 408 LLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGG 587
Query: 416 LLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAP 474
+++V+IPRNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAP
Sbjct: 588 VMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAP 767
Query: 475 RGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDE 534
RG I VCF ID +GILNVSA + T G + +ITITN+KGRLS E+IE+M+QEAE +K E
Sbjct: 768 RGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSE 947
Query: 535 DMKFKKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHE 594
D + KKKV+A N L++Y Y MR +KD+ + L ++ KI A+ + ++ +D Q E
Sbjct: 948 DEEHKKKVEAKNALENYAYNMRNTIKDEKIGGKLDPADKKKIEDAIEQAIQWLDSNQLAE 1127
Query: 595 A 595
A
Sbjct: 1128A 1130
Score = 28.5 bits (62), Expect(2) = e-122
Identities = 11/16 (68%), Positives = 14/16 (86%)
Frame = +1
Query: 221 KNNLFEVKATSGDTHL 236
+ +FEVKAT+GDTHL
Sbjct: 1 EEGIFEVKATAGDTHL 48
>TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (74%)
Length = 1752
Score = 312 bits (800), Expect(2) = 1e-89
Identities = 180/380 (47%), Positives = 242/380 (63%), Gaps = 5/380 (1%)
Frame = +3
Query: 135 EIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALAYGLQKR 194
E AE +L V AV+TVPAYFND+ R+ATKDAG IAGL+V RIINEPTAAAL+YG+ +
Sbjct: 3 ETAEAYLGKSVSKAVITVPAYFNDAHRQATKDAGRIAGLDVQRIINEPTAAALSYGMTNK 182
Query: 195 ANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNHFVMEFN 254
E I +FDLGGGTFDVS+L I N +FEVKAT+GDT LGGEDFDN +++ V EF
Sbjct: 183 -----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVDEFK 347
Query: 255 RKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FHSSITRAKF 310
R D+S L+RLR A E+ K LS + I++ + + ++TR+KF
Sbjct: 348 RTESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKF 527
Query: 311 EQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDICVS 370
E L L ++ ++CL DA I VD+ +LVGG +R+PKVQ+++ F K
Sbjct: 528 EALVNHLIERTKAPCKNCLKDANISIKEVDEVLLVGGMTRVPKVQEVVSAIFG-KSPSKG 704
Query: 371 INPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRNTPIPVK 430
+NPDEAVA GAA+Q +L ++ L+L DVTPLSLG G + + +I RNT IP K
Sbjct: 705 VNPDEAVAMGAAIQGGILRGDVKE---LLLLDVTPLSLGIETLGGIFTRLINRNTTIPTK 875
Query: 431 KTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIKVCFSIDFD 489
K++ + T D+ + V ++V +GER AS+N LG+F IPPAPRG I+V F ID +
Sbjct: 876 KSQVFSTAADNQTQVGIKVLQGEREMASDNKSLGEFELVGIPPAPRGLPQIEVTFDIDAN 1055
Query: 490 GILNVSATEDTCGNRQEITI 509
GI+ VSA + + G Q+ITI
Sbjct: 1056GIVTVSAKDKSTGKEQQITI 1115
Score = 35.0 bits (79), Expect(2) = 1e-89
Identities = 20/62 (32%), Positives = 33/62 (52%), Gaps = 3/62 (4%)
Frame = +2
Query: 514 GRLSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKVM---KDDSVSSMLTS 570
G LS +EIE+M++EAE +D + K + N D +Y++ K + +D S +
Sbjct: 1127 GGLSDDEIEKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKE 1306
Query: 571 IE 572
IE
Sbjct: 1307 IE 1312
>BP044919
Length = 519
Score = 272 bits (696), Expect = 1e-73
Identities = 136/164 (82%), Positives = 148/164 (89%)
Frame = +3
Query: 80 GRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSMVLTKMREIAEK 139
GRKYSDSVIQ D++LWPFKV+ G DDKP I+VK+KG EK EEISSMVL+KM+EIAE
Sbjct: 27 GRKYSDSVIQEDVKLWPFKVIAGTDDKPMIVVKHKGEEKKLVPEEISSMVLSKMKEIAEA 206
Query: 140 FLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALAYGLQKRANCVE 199
FLES VKNAVVTVPAYFND+QRKATKDAG IAGLNVMRIINEPTAAALAYGLQKRAN V
Sbjct: 207 FLESSVKNAVVTVPAYFNDAQRKATKDAGTIAGLNVMRIINEPTAAALAYGLQKRANLVG 386
Query: 200 ERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDN 243
ERN+FIFDLGGGTFDVSLLTIK++ FEVKATSGDTHLG +DFDN
Sbjct: 387 ERNVFIFDLGGGTFDVSLLTIKDDNFEVKATSGDTHLGXKDFDN 518
>BP040870
Length = 505
Score = 260 bits (665), Expect = 4e-70
Identities = 129/167 (77%), Positives = 144/167 (85%)
Frame = +2
Query: 111 VKYKGGEKHFCAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAI 170
V YKG EK F AEEISSMVL KM+EIAE +L + +KNAVVTVPAYFNDSQR+ATKDAG I
Sbjct: 5 VNYKGEEKQFSAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVI 184
Query: 171 AGLNVMRIINEPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKAT 230
+GLNVMRIINEPTAAA+AYGL K+A E+N+ IFDLGGGTFDVSLLTI+ +FEVK+T
Sbjct: 185 SGLNVMRIINEPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKST 364
Query: 231 SGDTHLGGEDFDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACER 277
+GDTHLGGEDFDNRMVNHFV EF RK KDISG+ R LRRLRTACER
Sbjct: 365 AGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACER 505
>BP041494
Length = 572
Score = 166 bits (419), Expect(2) = 4e-64
Identities = 82/110 (74%), Positives = 93/110 (84%)
Frame = +3
Query: 74 DAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSMVLTKM 133
DAKRLIGRK+SD +++D++LWPFKV+ G D+P I+V YK EK F AEEISSMVL KM
Sbjct: 243 DAKRLIGRKFSDDSVESDMKLWPFKVISGPVDRPMIVVNYKNEEKQFTAEEISSMVLIKM 422
Query: 134 REIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPT 183
REIAE +L S VKN VVTVPAYFNDSQR+ATKDAG IAGLNVMRIINEPT
Sbjct: 423 REIAEAYLGSTVKNVVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 572
Score = 96.7 bits (239), Expect(2) = 4e-64
Identities = 49/66 (74%), Positives = 54/66 (81%)
Frame = +1
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
M + + PAIGIDLGTTYSCV VWQ N+R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 61 MVGEGESPAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDFERLIGDAA 234
Query: 61 KNQAAS 66
KNQ S
Sbjct: 235 KNQMRS 252
>TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment), partial
(63%)
Length = 981
Score = 220 bits (561), Expect = 4e-58
Identities = 117/178 (65%), Positives = 136/178 (75%), Gaps = 1/178 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNR TPS+VAF D +RLIG+AAKNQ A
Sbjct: 145 GTVIGIDLGTTYSCVGVYR--NGHVEIIANDQGNRITPSWVAFNDGERLIGEAAKNQFAV 318
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGE-KHFCAEEI 125
NP TIFD KRLIGRK+ D +Q D++L P+K+V D KP I VK K GE K F EE+
Sbjct: 319 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPEEV 495
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPT 183
S+M+LTKM+E AE FL + +AVVTVPAYFND+QR+ATKDAG IAGLNV RIINEPT
Sbjct: 496 SAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 669
Score = 121 bits (303), Expect = 4e-28
Identities = 61/111 (54%), Positives = 78/111 (69%)
Frame = +2
Query: 173 LNVMRIINEPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSG 232
L ++ + P AAA+AYGL K+ E+NI +FDLGGGTFDVS+LTI N +FEV AT+G
Sbjct: 638 LMLLELSMNPPAAAIAYGLDKKGG---EKNILVFDLGGGTFDVSILTIDNGVFEVLATNG 808
Query: 233 DTHLGGEDFDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLS 283
DTHLGGEDFD R++ +F+ +K+ KDIS R L +LR ER KR LS
Sbjct: 809 DTHLGGEDFDQRIMEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALS 961
>TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (23%)
Length = 508
Score = 215 bits (548), Expect = 1e-56
Identities = 109/152 (71%), Positives = 126/152 (82%)
Frame = +2
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 59 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 232
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP+NT+FDAKRLIGR+ SD+ +Q+D++LWPFKV G +KP I V YKG +K F
Sbjct: 233 KNQVAMNPINTVFDAKRLIGRRVSDASVQSDMKLWPFKVNSGPAEKPMIQVSYKGEDKQF 412
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTV 152
AEEISSMVL KMREIAE +L S +KNAVVTV
Sbjct: 413 AAEEISSMVLMKMREIAEAYLGSTIKNAVVTV 508
>AW720237
Length = 459
Score = 197 bits (500), Expect = 5e-51
Identities = 96/141 (68%), Positives = 112/141 (79%)
Frame = +3
Query: 9 AIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNP 68
AIGIDLGTTYSCV VW N+R EII NDQGNRTTPS+V FT+ +RLIGDAAKNQ A NP
Sbjct: 42 AIGIDLGTTYSCVGVWM--NDRVEIIANDQGNRTTPSYVGFTESERLIGDAAKNQVARNP 215
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSM 128
+NT+FDAKRLIGRK+ D ++Q DI+LWPFKV G D KP I VKYKG K F AE++SSM
Sbjct: 216 INTVFDAKRLIGRKFDDPIVQKDIKLWPFKVEAGPDGKPLIAVKYKGETKKFHAEQVSSM 395
Query: 129 VLTKMREIAEKFLESPVKNAV 149
VLTKM+E AE +L +K+ V
Sbjct: 396 VLTKMKETAEAYLNKTIKDIV 458
>BP064075
Length = 554
Score = 173 bits (439), Expect = 6e-44
Identities = 86/141 (60%), Positives = 114/141 (79%), Gaps = 1/141 (0%)
Frame = -2
Query: 289 TIDIDSLFEGIDFHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGS 348
TI++D L++GIDF+SSITRAKFE+LN D F+KC+E VE C+ D++++KS++ D VLVGGS
Sbjct: 553 TIELDFLYQGIDFYSSITRAKFEELNKDYFQKCMELVEKCVIDSKMDKSNIHDVVLVGGS 374
Query: 349 SRIPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLS 407
SRIPKV+QLL +FF KD+C INPDEAVA+GAAV A++L E +EK +L+LR+VTPLS
Sbjct: 373 SRIPKVRQLLMDFFGRKDLCNRINPDEAVAHGAAVHASILSGEFSEKVQSLLLREVTPLS 194
Query: 408 LGTLVRGDLLSVVIPRNTPIP 428
LG G ++ +IPRNT IP
Sbjct: 193 LGLEKHGGIMETIIPRNTMIP 131
>TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, partial
(24%)
Length = 476
Score = 147 bits (372), Expect = 4e-36
Identities = 80/155 (51%), Positives = 110/155 (70%), Gaps = 2/155 (1%)
Frame = +3
Query: 346 GGSSRIPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVT 404
GGS+RIPKVQQLLK++F K+ +NPDEAVAYGAAVQ ++L E E++ +++L DV
Sbjct: 3 GGSTRIPKVQQLLKDYFEGKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVA 182
Query: 405 PLSLGTLVRGDLLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLG 464
PL+LG G +++ +IPRNT IP KK++ + T +D + VS++V+EGER + LLG
Sbjct: 183 PLTLGIETVGGVMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLG 362
Query: 465 KFRFS-IPPAPRGQIPIKVCFSIDFDGILNVSATE 498
KF S IPPAPRG I+V F +D +GILNV A +
Sbjct: 363 KFDLSGIPPAPRGTPQIEVTFEVDANGILNVKAED 467
>BP083571
Length = 423
Score = 132 bits (333), Expect = 1e-31
Identities = 61/72 (84%), Positives = 69/72 (95%)
Frame = +1
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K++GPAIGIDLGTTYSCVAVWQ+QN+RAEIIHN+QGNRTTPSFVAFTD +RLIGDAA
Sbjct: 145 MATKYEGPAIGIDLGTTYSCVAVWQEQNDRAEIIHNEQGNRTTPSFVAFTDTQRLIGDAA 324
Query: 61 KNQAASNPVNTI 72
KNQAA+NP NT+
Sbjct: 325 KNQAATNPTNTV 360
>AV766006
Length = 408
Score = 125 bits (315), Expect = 2e-29
Identities = 66/134 (49%), Positives = 90/134 (66%), Gaps = 1/134 (0%)
Frame = -1
Query: 414 GDLLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPP 472
G ++S +I RNT IP +KT+ + T +D + VS++VYEGER +N LGKF + IP
Sbjct: 405 GGVMSKIIERNTVIPTRKTQTFTTYQDKQTTVSIQVYEGERAMTKDNRQLGKFDLNGIPA 226
Query: 473 APRGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFK 532
A RG I+V F +D +GIL VSA + ++ ITITN+KGRLS EEIERM++EAE F
Sbjct: 225 AARGTPQIEVTFEVDANGILAVSAEDKASSAKEAITITNDKGRLSKEEIERMVKEAEEFA 46
Query: 533 DEDMKFKKKVKAMN 546
D D + K+KV+ N
Sbjct: 45 DADAEVKRKVEGKN 4
>AV769455
Length = 556
Score = 122 bits (305), Expect = 2e-28
Identities = 68/184 (36%), Positives = 102/184 (54%), Gaps = 1/184 (0%)
Frame = +1
Query: 56 IGDAAKNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKG 115
+G A + NP N+I KRLIGR+++D +Q D++ PF V G D P I +Y G
Sbjct: 4 LGTAGASSTMMNPKNSISQLKRLIGRQFADPELQRDLKSLPFAVTEGPDGFPLIHARYLG 183
Query: 116 GEKHFCAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNV 175
+ F ++ M+L+ ++EIA+K L + V + + +P YF D QR+A DA IAGL+
Sbjct: 184 ETRTFTPTQVFGMMLSNLKEIAQKNLNAAVVDCCIGIPIYFTDLQRRAVLDAATIAGLHP 363
Query: 176 MRIINEPTAAALAYGLQKRANCVEE-RNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDT 234
+R+ +E TA ALAYG+ K + N+ D+G + V + K +V A S D
Sbjct: 364 LRLFHETTATALAYGIYKTDLPENDPLNVAFVDVGHASMQVCIAGYKKGQLKVLAHSYDR 543
Query: 235 HLGG 238
LGG
Sbjct: 544 SLGG 555
>TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (48%)
Length = 746
Score = 112 bits (279), Expect = 2e-25
Identities = 61/143 (42%), Positives = 90/143 (62%), Gaps = 3/143 (2%)
Frame = +1
Query: 440 DDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKVCFSIDFDGILNVSATE 498
D+ + V ++VYEGE KA +N LGKF S PAP+G I VCF +D DGI+ VSA +
Sbjct: 1 DNQTSVWIKVYEGEGAKADKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED 180
Query: 499 DTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKV 558
+ ++ ITITN GRLS EE+ RM+++A +K ED + +KKVKA N L++Y Y R
Sbjct: 181 MSLRLKKTITITNRLGRLSQEEMSRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDR 360
Query: 559 MK--DDSVSSMLTSIERMKINSA 579
+K + V ++ ++R ++ A
Sbjct: 361 VKKLEKMVEEVIEWLDRNQLAEA 429
>BP033771
Length = 513
Score = 111 bits (278), Expect = 3e-25
Identities = 56/116 (48%), Positives = 77/116 (66%)
Frame = -2
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
P+ VCF +D +GILNVSA E+T G + IT+TN+ GRLS+E+I R+IQEAE + ED +
Sbjct: 512 PLNVCFDLDANGILNVSAEEETTGIKNGITVTNDNGRLSSEQIMRLIQEAEKHRAEDRMY 333
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHE 594
+KKV+AMN LDDY+Y +R +KD SS L ++ KI A+ K L+ E
Sbjct: 332 EKKVEAMNALDDYVYKLRNAIKDIDRSSKLRPQDKSKIRVAVTKARNLLGTSDQTE 165
>AW163930
Length = 359
Score = 101 bits (252), Expect = 3e-22
Identities = 54/93 (58%), Positives = 68/93 (73%), Gaps = 1/93 (1%)
Frame = +1
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++V
Sbjct: 40 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 219
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEG 452
+IPRNT IP KK + + T D+ V ++VYEG
Sbjct: 220 LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEG 318
>CB827219
Length = 544
Score = 85.5 bits (210), Expect = 2e-17
Identities = 51/177 (28%), Positives = 90/177 (50%)
Frame = +3
Query: 191 LQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNHFV 250
L R V R + F++G G DV++ + ++KA +G T +GGED M++H +
Sbjct: 6 LLMRIWAVGVRKLLYFNMGAGYCDVAVTVTAGGVSQIKALAGST-IGGEDLLQNMMHHLL 182
Query: 251 MEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRAKF 310
+ + + + LR A LS + +D+D L +G+ + I +F
Sbjct: 183 PDCENIFKNHGVKEIKSMGLLRVATLDAIHRLSSQKSVQVDVD-LGDGLKINKVIDCEEF 359
Query: 311 EQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDI 367
E++N ++F+KC + CL D++IE V+D ++VGG S IP+V+ L+ K +
Sbjct: 360 EEVNKNVFEKCESLIIQCLQDSKIEVEDVNDVIIVGGCSYIPRVKNLVTNICKGKKL 530
>AW428869
Length = 181
Score = 73.2 bits (178), Expect = 1e-13
Identities = 32/53 (60%), Positives = 42/53 (78%)
Frame = +1
Query: 203 IFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNHFVMEFNR 255
I +FDLGGGTFDVS+L + + +FEV +TSGDTHLGG+DFD R+V+ +F R
Sbjct: 22 ILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRIVDWLAADFKR 180
>BP058182
Length = 449
Score = 72.8 bits (177), Expect = 2e-13
Identities = 38/66 (57%), Positives = 47/66 (70%)
Frame = -3
Query: 529 ENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLID 588
E+ + +DMKFK KV AMNDLD YLY M K MKD VSS L S+ + KINSA++KG L+D
Sbjct: 447 EDHEVDDMKFKNKVTAMNDLDLYLYDMNKSMKDKCVSSKLPSMVQDKINSAIVKGXSLLD 268
Query: 589 EKQHHE 594
+ Q E
Sbjct: 267 DGQQLE 250
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,433,293
Number of Sequences: 28460
Number of extensions: 97015
Number of successful extensions: 440
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 416
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 418
length of query: 600
length of database: 4,897,600
effective HSP length: 95
effective length of query: 505
effective length of database: 2,193,900
effective search space: 1107919500
effective search space used: 1107919500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144722.13


