

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.7 + phase: 0 /pseudo
(817 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
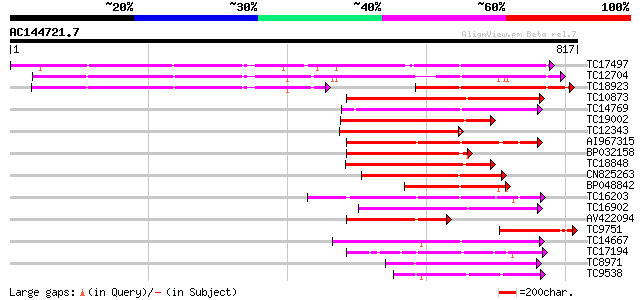
Score E
Sequences producing significant alignments: (bits) Value
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 519 e-148
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 507 e-144
TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1,... 242 2e-64
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 224 3e-59
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 213 1e-55
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 196 2e-50
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 193 1e-49
AI967315 193 1e-49
BP032158 190 9e-49
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 190 9e-49
CN825263 190 9e-49
BP048842 188 3e-48
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 187 4e-48
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 178 3e-45
AV422094 172 2e-43
TC9751 weakly similar to UP|O23743 (O23743) SFR1 protein precurs... 172 2e-43
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 171 3e-43
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 170 1e-42
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 160 1e-39
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 156 1e-38
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 519 bits (1336), Expect = e-148
Identities = 322/842 (38%), Positives = 470/842 (55%), Gaps = 57/842 (6%)
Frame = +1
Query: 1 MAFLSHNSNYFFIITFLIFCTIYSCYSAINDTITSSKSLKDN------------------ 42
+A + H+ N+ F++ +F +I +S + S +L+D+
Sbjct: 67 VATVRHHRNHLFLLRSAVFHSIIFSFSYVVCNGNGSLTLEDSSWKSGNYRCSSKKYEQCT 246
Query: 43 --ETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKTNNIWIANRDQPLKDSNG--IVTI 96
+ +T ++ F+ GFF N + Y G+WY I+ +W+ANRD PL++S +
Sbjct: 247 LSQGMTVHDGTFEAGFFHFENPQHHYFGVWYKSISPRTIVWVANRDAPLRNSTAPTLKVT 426
Query: 97 HKDGNFIILNKPNGVIIWSTNISSSTNST-AQLADSGNLILRDISSGAT-IWDSFTHPAD 154
HK G+ +I + GVI WSTN S + QL DSGNL+ +D G IW+SF +P D
Sbjct: 427 HK-GSILIRDGAKGVI-WSTNTSRAKEQPFMQLLDSGNLVAKDGDKGENVIWESFNYPGD 600
Query: 155 AAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGP 214
+ M+I +N G S ++ DP+SG +S ++ P++ + K I R GP
Sbjct: 601 TFLAGMKIKSNLAIGPTSYLTSWRNSEDPASGEFSYHIDIRGFPQLVVTKGAAITLRAGP 780
Query: 215 WNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMN 274
W G F G+ + + + + F Q TD + Y ++++ +TP GT++ + +
Sbjct: 781 WTGNKFSGAFGQVLQKILTF-FMQFTDQEISLEYETVNRSIITREVITPLGTIQRLLWSV 957
Query: 275 KKELFRLEVDQ--NECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCV 332
+ + + + + ++C Y CG CD S PIC C +GF P+ +W+ +W GCV
Sbjct: 958 RNQSWEIIATRPVDQCADYVFCGANSLCDTSKNPICDCLEGFMPQFQAKWNSLDWAGGCV 1137
Query: 333 RKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVRT--NNADQDKCGADCLANCSCL 390
E L C+ NG DGF + +K PD + N D+C CL NCSC
Sbjct: 1138SME--KLSCQ---NG------DGFMKHTGVKLPDTSSSWFGKNMSLDECRTLCLQNCSCT 1284
Query: 391 AYA-----YDPSIFCMYWTGELIDLQKFPNG--GVDLFVRVPAELVAVKKEKGHNKSF-- 441
AYA D S+ C+ W G+++D+ K P+ G ++++RV VA K ++ NK
Sbjct: 1285AYAGLDNDVDRSV-CLIWFGDILDMSKHPDPDQGQEIYIRV----VASKLDRTRNKKSIN 1449
Query: 442 -------LIIVIAGVIGALILVIC-AYLLWRKCSAR----------HKAREHQQMKLDEL 483
L+++IA VI IL + + + RK + R H + +D
Sbjct: 1450TKKLAGSLVVIIAFVIFITILGLAISTCIQRKKNKRGDEGEIGIINHWKDKRGDEDIDLA 1629
Query: 484 PLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEV 543
++DF + +ATN F +N LG+GGFGPVYKG++ +GQEIAVKRLS SGQG+EEF NE+
Sbjct: 1630TIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEI 1809
Query: 544 VVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEG 603
+I++LQHRNLV+L GC V + E NK + L D + K +DW KR II+G
Sbjct: 1810KLIARLQHRNLVKLFGCSVHQDENSHA-----NKKMK-ILLDSTRSKLVDWNKRLQIIDG 1971
Query: 604 IARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVG 663
IARG++YLH+DSRLRIIHRDLK SNILLD +M PKISDFGLARI G+ EA TKRV+G
Sbjct: 1972IARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIF-IGDQVEARTKRVMG 2148
Query: 664 TYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEE 723
TYGYMPPEYA+ G FS KSDV+SFGV++LEI+SG++ F L+L+ AW+LW+EE
Sbjct: 2149TYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEE 2328
Query: 724 NIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRV 783
+ L+D + D + +LR IH+ LLCVQ P +RP++ ++VLML E LP P
Sbjct: 2329RPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGE-KELPKPRLP 2505
Query: 784 AF 785
AF
Sbjct: 2506AF 2511
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 507 bits (1305), Expect = e-144
Identities = 313/828 (37%), Positives = 454/828 (54%), Gaps = 59/828 (7%)
Frame = +1
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKTNNIWIANRDQPLKDS 90
+ +S++D+ET+ S F+ GFF NS RY GIWY I+ +W+ANRD P+++S
Sbjct: 1 MAQKQSIQDDETLVSPEGTFEAGFFRFGNSLRRYFGIWYKSISPRTIVWVANRDAPVQNS 180
Query: 91 NGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNSTA-QLADSGNLILRDISSGAT-IWDS 148
+ + GN +IL+ G I+WS+N S + + QL DSGN +++D IW+S
Sbjct: 181 TATLKLTDQGNLLILDGLKG-IVWSSNASRTKDKPLMQLLDSGNFVVKDGDKEENLIWES 357
Query: 149 FTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFIWKDKNI 208
F +P D + M+I +N TG S ++ DP+SG +S ++ P++ + K +
Sbjct: 358 FDYPGDTFLAGMKIKSNLATGPTSYLTSWRNAEDPASGEFSYHIDTHGYPQLVVTKGATV 537
Query: 209 HWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADKTMFGILSLTPHGTLK 268
R GPW G F G+ + + + + Q TD + Y A++++ +TP GT +
Sbjct: 538 TLRAGPWIGNKFSGASGLRLQKILTFSM-QFTDKEVSLEYETANRSIITRTVITPSGTTQ 714
Query: 269 LIEYMNKKELFRL--EVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGN 326
+ + ++ + + + ++C +Y CG CD S PIC C +GF PK +W+ +
Sbjct: 715 RLLWSDRSQSWEIISTHPMDQCAYYAFCGANSMCDTSNNPICDCLEGFTPKFQAQWNSLD 894
Query: 327 WTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVRT--NNADQDKCGADCL 384
WT GCV + NL C+ NG DGF + ++ PD + N+ D+CG CL
Sbjct: 895 WTGGCVPIK--NLSCQ---NG------DGFPKHTGVQFPDTSSSWYGNSKSLDECGTICL 1041
Query: 385 ANCSCLAYAYDPSI----FCMYWTGELIDLQKFPNG--GVDLFVRVPAELVAVKKEKGHN 438
NCSC AYAY ++ C+ W G+++D+ + P+ G ++++RV A + ++ K
Sbjct: 1042QNCSCTAYAYLDNVGGRSVCLNWFGDILDMSEHPDPDQGQEIYLRVVASELDHRRNK--- 1212
Query: 439 KSFLIIVIAGVI-GALILVICAYLLW--------RKCSAR------------HKAREHQQ 477
KS I +AG + G++ +IC +L RK + R H +
Sbjct: 1213KSINIKKLAGSLAGSIAFIICITILGLATVTCIRRKKNEREDEGGIETRIINHWKDKRGD 1392
Query: 478 MKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIE 537
+D ++DF + + TN F +N LG+GGFGPVYKGV+ +GQEIAVKRLS SGQG+E
Sbjct: 1393EDIDLATIFDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTSGQGME 1572
Query: 538 EFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKR 597
EF NEV +I++LQHRNLV+LLGC + E +L+YEFM N+SLD F+F
Sbjct: 1573EFKNEVKLIARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIF------------- 1713
Query: 598 SNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEAN 657
DSRLRIIHRDLK SNILLDS+M PKISDFGLARI G+ EA
Sbjct: 1714----------------DSRLRIIHRDLKTSNILLDSEMNPKISDFGLARIFT-GDQVEAK 1842
Query: 658 TKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSF---SHHEDTLS--- 711
TKRV+GTYGYM PEYA+ G FS KSDV+SFGV++LEI+SG++ F HH + LS
Sbjct: 1843TKRVMGTYGYMSPEYAVHGSFSVKSDVFSFGVIVLEIISGKKIGRFCDPHHHRNLLSHSS 2022
Query: 712 -----------LVGF-------AWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCV 753
+ F AW+LW+EE + L+D + + +LR IHI LLCV
Sbjct: 2023NFAVFLIKALRICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTEILRYIHIALLCV 2202
Query: 754 QELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKS 801
Q+ P RP++ +VVLML E LP P AF ES+ K+
Sbjct: 2203QQRPEYRPDMLSVVLMLNGE-KELPKPSLPAFYTGNDDLLWPESTSKN 2343
>TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1, complete
Length = 2061
Score = 242 bits (618), Expect = 2e-64
Identities = 148/447 (33%), Positives = 239/447 (53%), Gaps = 16/447 (3%)
Frame = +1
Query: 32 TITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKTNNIWIANRDQPLKD 89
T+T ++S++D+ET+ S F+ GFF NS +Y GIWY I+ +W+ANRD P+++
Sbjct: 31 TVTQNQSIQDDETLVSAEGTFEAGFFGLGNSQRQYFGIWYKSISPRTIVWVANRDAPVQN 210
Query: 90 SNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNST-AQLADSGNLILRDISSGAT--IW 146
S + + GN +IL+ G IIWS+N S + QL DSGNL+++D IW
Sbjct: 211 STATIKLTDKGNLLILDGSKG-IIWSSNGSRAAEKPYMQLLDSGNLVVKDGGKRKKNLIW 387
Query: 147 DSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFIWKDK 206
+SF +P D + M+I +N V G S ++ DP+SG +S ++ P++ I ++
Sbjct: 388 ESFDYPGDTLLAGMKIKSNLVKGPTSYLTSWRNTEDPASGEFSYLIDTRGFPQLVITRNA 567
Query: 207 NIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADKTMFGILSLTPHGT 266
++R GPW G++F GS + + + Q T + Y A++++ + P GT
Sbjct: 568 TAYYRAGPWTGKLFSGSSWLRLRKILTFSM-QFTSQEISLEYETANRSIITRAVINPSGT 744
Query: 267 LKLIEYMNKKELFRL--EVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSL 324
+ + + ++ + + + ++C +YG CG CD S PIC C +GF PK +W+
Sbjct: 745 TQRLLWSDRSQSWEIISTHPTDQCTYYGLCGANSMCDISNNPICHCLEGFRPKFQAKWNS 924
Query: 325 GNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVR--TNNADQDKCGAD 382
+W GCV + NL C+ NG DGF + +K PD + N D+CG
Sbjct: 925 FDWPGGCVPMK--NLSCQ---NG------DGFLKHTGVKLPDTSSSWYGKNKSLDECGTL 1071
Query: 383 CLANCSCLAYAY-DPSI---FCMYWTGELIDLQKFPN--GGVDLFVRVPAELVAVKKEKG 436
CL NCSC +YAY D I C+ W G+++DL PN G +++++V A + ++ K
Sbjct: 1072CLQNCSCTSYAYLDNDIGGSACLIWFGDILDLSIHPNPDQGQEIYIKVVASELDHRRNK- 1248
Query: 437 HNKSFLIIVIAG-VIGALILVICAYLL 462
KSF+ +AG + G + LVIC +L
Sbjct: 1249--KSFMTKKLAGSLAGIVALVICIIIL 1323
Score = 223 bits (567), Expect = 1e-58
Identities = 119/229 (51%), Positives = 155/229 (66%)
Frame = +1
Query: 585 DPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGL 644
D + K LDW KR II+GIARG++YLH+DSRLRIIHRDLK SNILLD++M PKISDFGL
Sbjct: 1378 DSTRSKLLDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDNEMNPKISDFGL 1557
Query: 645 ARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFS 704
ARI G+ EA TKRV+GTYGYMPPEYA+ G FS KSDV+SFGV++LEI+SG++ F
Sbjct: 1558 ARIF-IGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIRKFY 1734
Query: 705 HHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNIS 764
L+L+ AW+LW+E + + L+D D+ + +LR IH+ LLCVQ P RP++
Sbjct: 1735 DPHHHLNLLSHAWRLWIEGSPLELVDKLFEDSIIPTEILRYIHVALLCVQRRPETRPDML 1914
Query: 765 TVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLS 813
++VLML E LP P AF + ES + + +VT+S
Sbjct: 1915 SIVLMLNGE-KELPKPSLPAFYTGKHDPILLESPSR----RCSTSVTIS 2046
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 224 bits (572), Expect = 3e-59
Identities = 122/287 (42%), Positives = 183/287 (63%), Gaps = 2/287 (0%)
Frame = +1
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVV 545
+ F +L AT F N++G+GGFG VYKG + G+ +AVK+LS QG +EF+ EV++
Sbjct: 415 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLM 594
Query: 546 ISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFD-PLQKKNLDWRKRSNIIEGI 604
+S L H NLVRL+G C + +++LVYE+MP SL+ LF+ K+ L+W R + G
Sbjct: 595 LSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGA 774
Query: 605 ARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGT 664
ARG+ YLH + +I+RDLK++NILLD++ PK+SDFGLA++ G++ +T RV+GT
Sbjct: 775 ARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVST-RVMGT 951
Query: 665 YGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEEN 724
YGY PEYAM G + KSD+YSFGV+LLE+++GRR S +LV +A + +
Sbjct: 952 YGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRR 1131
Query: 725 IIS-LIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
++DP + + + I I +C+QE P+ RP I+ +V+ L
Sbjct: 1132RFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVAL 1272
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 213 bits (541), Expect = 1e-55
Identities = 121/291 (41%), Positives = 168/291 (57%), Gaps = 1/291 (0%)
Frame = +3
Query: 478 MKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIE 537
+K + + E +E AT + ++G+GGFG VY+G + DGQE+AVK S S QG
Sbjct: 2109 IKSVSIQAFTLEYIEVATE--RYKTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTR 2282
Query: 538 EFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLF-DPLQKKNLDWRK 596
EF NE+ ++S +QH NLV LLG C E +QILVY FM N SL L+ +P ++K LDW
Sbjct: 2283 EFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPT 2462
Query: 597 RSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEA 656
R +I G ARG+ YLH +IHRD+K+SNILLD M K++DFG ++ E D
Sbjct: 2463 RLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAP-QEGDSY 2639
Query: 657 NTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFA 716
+ V GT GY+ PEY SEKSDV+SFGV+LLEIVSGR + SLV +A
Sbjct: 2640 VSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWA 2819
Query: 717 WKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVV 767
+ ++DP + +M R + + L C++ RP++ +V
Sbjct: 2820 TPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIV 2972
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 196 bits (497), Expect = 2e-50
Identities = 103/224 (45%), Positives = 148/224 (65%), Gaps = 1/224 (0%)
Frame = +1
Query: 477 QMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGI 536
Q K ++ ++L +ATN F+++N LG+GGFG VY G + DG +IAVKRL S +
Sbjct: 1 QKKQPAWRVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKAD 180
Query: 537 EEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKK-NLDWR 595
EF EV ++++++H+NL+ L G C E E+++VY++MPN SL + L + LDW
Sbjct: 181 MEFAVEVEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWN 360
Query: 596 KRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDE 655
+R NI G A GI+YLH + IIHRD+KASN+LLDSD +++DFG A+++ G
Sbjct: 361 RRMNIAIGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHV 540
Query: 656 ANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
T RV GT GY+ PEYAM G +E DV+SFG+LLLE+ SG++
Sbjct: 541 --TTRVKGTLGYLAPEYAMLGKANECCDVFSFGILLLELASGKK 666
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 193 bits (490), Expect = 1e-49
Identities = 94/178 (52%), Positives = 128/178 (71%)
Frame = +3
Query: 476 QQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQG 535
Q + E + +E L AT FH N LG+GGFGPV+KG + DG+EIAVK+LS+ S QG
Sbjct: 177 QNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQG 356
Query: 536 IEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWR 595
+F+NE +++++QHRN+V L G C E++LVYE++P +SLD LF +K+ LDW+
Sbjct: 357 RTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQLDWK 536
Query: 596 KRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGED 653
+R +II G+ARG++YLH DS IIHRD+KA+NILLD +PKI+DFGLARI F ED
Sbjct: 537 RRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARI--FPED 704
>AI967315
Length = 1308
Score = 193 bits (490), Expect = 1e-49
Identities = 114/285 (40%), Positives = 171/285 (60%), Gaps = 3/285 (1%)
Frame = +1
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKA--SGQGIEEFMNEV 543
+ +E+L ATN F NM+GKGG+ VYKG +E G EIAVKRL++ + +EF+ E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 544 VVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEG 603
I + H N++ LLGCC++ G LV+E S+ + + D + LDW+ R I+ G
Sbjct: 292 GTIGHVCHSNVMPLLGCCIDNG-LYLVFELSTVGSVASLIHDE-KMAPLDWKTRYKIVLG 465
Query: 604 IARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVG 663
ARG+ YLH+ + RIIHRD+KASNILL D P+ISDFGLA+ + + + + G
Sbjct: 466 TARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLP-SQWTHHSIAPIEG 642
Query: 664 TYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEE 723
T+G++ PEY M G+ EK+DV++FGV LLE++SGR+ SH SL +A + +
Sbjct: 643 TFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSHQ----SLHTWAKPILSKW 810
Query: 724 NIISLIDPEVWDACFESSML-RCIHIGLLCVQELPRDRPNISTVV 767
I L+DP + + C++ + R LC++ RP +S V+
Sbjct: 811 EIEKLVDPRL-EGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVL 942
>BP032158
Length = 555
Score = 190 bits (482), Expect = 9e-49
Identities = 98/185 (52%), Positives = 130/185 (69%), Gaps = 4/185 (2%)
Frame = +2
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVV 545
+ +++ ATN F N +G+GGFGPVYKGV+ +G IAVK+LS S QG EF+NE+ +
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFINEIGM 193
Query: 546 ISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLF-DPLQKKNLDWRKRSNIIEGI 604
IS LQH NLV+L GCC+E + +LVYE+M N SL LF + QK NL+WR R I GI
Sbjct: 194 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVGI 373
Query: 605 ARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANT---KRV 661
A+G+ YLH +SRL+I+HRD+KA+N+LLD D+ KISDFGLA++ D+E NT R+
Sbjct: 374 AKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKL-----DEEENTHISTRI 538
Query: 662 VGTYG 666
GT G
Sbjct: 539 AGTIG 553
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 190 bits (482), Expect = 9e-49
Identities = 100/216 (46%), Positives = 142/216 (65%), Gaps = 1/216 (0%)
Frame = +2
Query: 485 LYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVV 544
++ +++L AT F +N LG+GGFG VY G DG +IAVK+L + + EF EV
Sbjct: 263 IFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVE 442
Query: 545 VISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQ-KKNLDWRKRSNIIEG 603
V+ +++H+NL+ L G CV ++++VY++MPN SL + L + L+W+KR I G
Sbjct: 443 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIG 622
Query: 604 IARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVG 663
A GI+YLH + IIHRD+KASN+LL+SD P ++DFG A+++ G T RV G
Sbjct: 623 SAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSH--MTTRVKG 796
Query: 664 TYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
T GY+ PEYAM G SE DVYSFG+LLLE+V+GR+
Sbjct: 797 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRK 904
>CN825263
Length = 663
Score = 190 bits (482), Expect = 9e-49
Identities = 98/210 (46%), Positives = 140/210 (66%), Gaps = 1/210 (0%)
Frame = +1
Query: 508 GFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQ 567
GFG VYKG++ DG+++AVK L + +G EF+ EV ++S+L HRNLV+L+G C+E+ +
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 568 ILVYEFMPNKSLDAFLFDPLQKKN-LDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKA 626
L+YE +PN S+++ L ++ LDW R I G ARG+ YLH DS +IHRD K+
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 627 SNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYS 686
SNILL+ D PK+SDFGLAR E ++ + V+GT+GY+ PEYAM G KSDVYS
Sbjct: 361 SNILLECDFTPKVSDFGLAR-TALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 687 FGVLLLEIVSGRRNSSFSHHEDTLSLVGFA 716
+GV+LLE+++G + S +LV +A
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQENLVTWA 627
>BP048842
Length = 524
Score = 188 bits (477), Expect = 3e-48
Identities = 99/159 (62%), Positives = 121/159 (75%), Gaps = 7/159 (4%)
Frame = -3
Query: 570 VYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNI 629
+YEFM N+SL+ F+FD + K +DW KR II+GIARG++YLH+DSRLRIIHRDLK SNI
Sbjct: 522 IYEFMHNRSLNYFIFDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNI 343
Query: 630 LLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGV 689
LLD +M PKISDFGLARI G+ EA TKRV+GTYGYMPPEYA+ G FS KSDV+SFGV
Sbjct: 342 LLDDEMNPKISDFGLARIF-IGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGV 166
Query: 690 LLLEIVSGRRNSSF---SHHEDTLSLVG----FAWKLWL 721
++LEI+SG++ F HH + LS V F KL+L
Sbjct: 165 IVLEIISGKKIGRFYDPHHHLNLLSHVSLTCIFLSKLYL 49
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 187 bits (476), Expect = 4e-48
Identities = 115/351 (32%), Positives = 190/351 (53%), Gaps = 8/351 (2%)
Frame = +2
Query: 430 AVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDELPLYDFE 489
+++K + IVI + +L++ + + H+A+ + L +
Sbjct: 2000 SLRKTRAKTARVRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQRLEI---- 2167
Query: 490 KLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRL-SKASGQGIEEFMNEVVVISK 548
K E C N++GKGG G VY+G M +G ++A+KRL + SG+ F E+ + K
Sbjct: 2168 KAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGK 2347
Query: 549 LQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGI 608
++HRN++RLLG + +L+YE+MPN SL +L + +L W R I ARG+
Sbjct: 2348 IRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGA-KGGHLRWEMRYKIAVEAARGL 2524
Query: 609 MYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYM 668
Y+H D IIHRD+K++NILLD+D ++DFGLA+ + + + + G+YGY+
Sbjct: 2525 CYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFL-YDPGASQSMSSIAGSYGYI 2701
Query: 669 PPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEEN---- 724
PEYA EKSDVYSFGV+LLE++ GR+ D + +VG+ K E +
Sbjct: 2702 APEYAYTLKVDEKSDVYSFGVVLLELIIGRK--PVGEFGDGVDIVGWVNKTMSELSQPSD 2875
Query: 725 ---IISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVS 772
+++++DP + +S++ +I ++CV+E+ RP + VV ML +
Sbjct: 2876 TALVLAVVDPRL-SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTN 3025
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 178 bits (451), Expect = 3e-45
Identities = 100/265 (37%), Positives = 156/265 (58%)
Frame = +3
Query: 503 MLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCV 562
+LG GGFG VY G ++ G ++A+KR + S QG+ EF E+ ++SKL+HR+LV L+G C
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 563 ERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHR 622
E E ILVY+ M +L L+ QK L W++R I G ARG+ YLH ++ IIHR
Sbjct: 192 ENTEMILVYDHMAYGTLREHLY-KTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHR 368
Query: 623 DLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKS 682
D+K +NILLD + K+SDFGL++ ++ +T V G++GY+ PEY ++KS
Sbjct: 369 DVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTV-VKGSFGYLDPEYFRRQQLTDKS 545
Query: 683 DVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSM 742
DVYSFGV+L EI+ R + S ++ +SL +A + + + ++DP +
Sbjct: 546 DVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIAPECF 725
Query: 743 LRCIHIGLLCVQELPRDRPNISTVV 767
+ + CV + +RP++ V+
Sbjct: 726 KKFAETAMKCVSDQGIERPSMGDVL 800
>AV422094
Length = 462
Score = 172 bits (436), Expect = 2e-43
Identities = 81/151 (53%), Positives = 114/151 (74%)
Frame = +2
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVV 545
+ + +L+ ATN F+ +N LG+GGFGPVYKG++ DG IAVK+LS S QG +F+ E+
Sbjct: 14 FSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTVIAVKQLSLGSHQGKSQFIAEIAT 193
Query: 546 ISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIA 605
IS +QHRNLV+L GCC+E +++LVYE++ NKSLD LF + +L+W R +I G+A
Sbjct: 194 ISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFG--KALSLNWSTRYDICLGVA 367
Query: 606 RGIMYLHRDSRLRIIHRDLKASNILLDSDMI 636
RG+ YLH +SRLRI+HRD+KASNILLD +++
Sbjct: 368 RGLTYLHEESRLRIVHRDVKASNILLDHELV 460
>TC9751 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (5%)
Length = 550
Score = 172 bits (436), Expect = 2e-43
Identities = 86/112 (76%), Positives = 100/112 (88%)
Frame = +1
Query: 706 HEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNIST 765
++D+LSL+GFAWKLW EENIISLIDPE+WD ESSMLRCIHIGLLCVQE+PR+RP IST
Sbjct: 7 NDDSLSLLGFAWKLWTEENIISLIDPEIWDPSVESSMLRCIHIGLLCVQEIPRERPAIST 186
Query: 766 VVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
VVLML+SEIT LPPPG+VAFV K ++KS ++SSQKS Q +SNNNVTLSEV G
Sbjct: 187 VVLMLISEITQLPPPGQVAFVQKHNAKS-SDSSQKS-QCSSNNNVTLSEVHG 336
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 171 bits (434), Expect = 3e-43
Identities = 109/313 (34%), Positives = 166/313 (52%), Gaps = 7/313 (2%)
Frame = +2
Query: 465 KCSARHKAREHQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIA 524
K SA K + E+P ++L+ T+ F ++G+G +G VY + DG +A
Sbjct: 260 KASAPVKHETQKAPPPIEVPALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVA 439
Query: 525 VKRLSKASG-QGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFL 583
VK+L +S + EF+ +V ++S+L++ N V L G CVE ++L YEF SL L
Sbjct: 440 VKKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDIL 619
Query: 584 FDPLQKKN------LDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIP 637
+ LDW +R I ARG+ YLH + IIHRD+++SN+L+ D
Sbjct: 620 HGRKGVQGAQPGPTLDWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKA 799
Query: 638 KISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSG 697
KI+DF L+ ++ RV+GT+GY PEYAM G ++KSDVYSFGV+LLE+++G
Sbjct: 800 KIADFNLSNQAP-DMAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTG 976
Query: 698 RRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELP 757
R+ + SLV +A E+ + +DP++ + + + LCVQ
Sbjct: 977 RKPVDHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEA 1156
Query: 758 RDRPNISTVVLML 770
RPN+S VV L
Sbjct: 1157EFRPNMSIVVKAL 1195
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 170 bits (430), Expect = 1e-42
Identities = 108/300 (36%), Positives = 172/300 (57%), Gaps = 10/300 (3%)
Frame = +1
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVV 545
+ +++L ATN F +N +G+GGFG VY + G++ A+K++ Q EF+ E+ V
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDV---QASTEFLCELKV 1218
Query: 546 ISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIA 605
++ + H NLVRL+G CVE G LVYE + N +L +L K+ L W R I A
Sbjct: 1219 LTHVHHLNLVRLIGYCVE-GSLFLVYEHIDNGNLGQYLHGS-GKEPLPWSSRVQIALDAA 1392
Query: 606 RGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTY 665
RG+ Y+H + IHRD+K++NIL+D ++ K++DFGL ++++ G + R+VGT+
Sbjct: 1393 RGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVG--NSTLQTRLVGTF 1566
Query: 666 GYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLE--- 722
GYMPPEYA G S K DVY+FGV+L E++S +N+ E G L+ E
Sbjct: 1567 GYMPPEYAQYGDISPKIDVYAFGVVLFELISA-KNAVLKTGELVAESKGLV-ALFEEALN 1740
Query: 723 -----ENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVV--LMLVSEIT 775
+ + L+DP + + S+L+ +G C ++ P RP++ ++V LM +S +T
Sbjct: 1741 KSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTLSSLT 1920
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 160 bits (404), Expect = 1e-39
Identities = 87/225 (38%), Positives = 136/225 (59%)
Frame = +3
Query: 542 EVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNII 601
EV +++K+ HRNLV+LLG + E+IL+ E++PN +L L D L+ K LD+ +R I
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHL-DGLRGKILDFNQRLEIA 182
Query: 602 EGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRV 661
+A G+ YLH + +IIHRD+K+SNILL M K++DFG AR+ D + +V
Sbjct: 183 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTKV 362
Query: 662 VGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWL 721
GT GY+ PEY + KSDVYSFG+LLLEI++GRR D + +A++ +
Sbjct: 363 KGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKYN 542
Query: 722 EENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTV 766
E +++ L+DP + +A +++ + + C + DRPN+ +V
Sbjct: 543 EGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSV 677
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 156 bits (394), Expect = 1e-38
Identities = 92/234 (39%), Positives = 135/234 (57%), Gaps = 14/234 (5%)
Frame = +2
Query: 553 NLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKN------------LDWRKRSNI 600
N+VRLL C +LVYE++ N SLD +L L+ K+ LDW KR I
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLH--LKPKSSSVSGVVQQYTVLDWPKRLKI 178
Query: 601 IEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLAR-IVKFGEDDEANTK 659
G A+G+ Y+H D I+HRD+K SNILLD K++DFGLAR ++K GE + +T
Sbjct: 179 AIGAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMST- 355
Query: 660 RVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWK- 718
V+GT+GY+ PEY SEK DVYSFGV+LLE+ +G+ + H SL +AW+
Sbjct: 356 -VIGTFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKEANYGDQHS---SLAEWAWRH 523
Query: 719 LWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVS 772
+ + N+ L+ +V +A + M +G++C LP RP++ V+ +L+S
Sbjct: 524 ILIGSNVXDLLXKDVMEASYIDEMCSVFKLGVMCTATLPATRPSMKEVLPILLS 685
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,023,054
Number of Sequences: 28460
Number of extensions: 244116
Number of successful extensions: 1793
Number of sequences better than 10.0: 337
Number of HSP's better than 10.0 without gapping: 1568
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1583
length of query: 817
length of database: 4,897,600
effective HSP length: 98
effective length of query: 719
effective length of database: 2,108,520
effective search space: 1516025880
effective search space used: 1516025880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144721.7


