

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
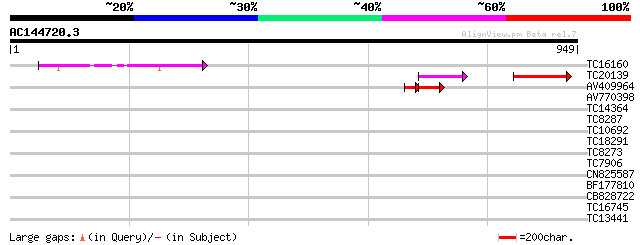
Query= AC144720.3 + phase: 0 /pseudo
(949 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16160 168 4e-42
TC20139 similar to GB|AAG31805.1|11139702|AF315590 Pumilio 2 {Mu... 148 5e-36
AV409964 66 1e-15
AV770398 38 0.007
TC14364 weakly similar to UP|Q9SVZ7 (Q9SVZ7) Pumilio-like protei... 36 0.033
TC8287 UP|AAQ87022 (AAQ87022) VDAC2.1, complete 32 0.36
TC10692 weakly similar to UP|BAD08916 (BAD08916) Calcium-binding... 32 0.62
TC18291 similar to UP|TRMU_BORBU (O51625) Probable tRNA (5-methy... 30 2.3
TC8273 UP|BAD11134 (BAD11134) Glutamate-rich protein, complete 29 3.1
TC7906 homologue to PIR|S46534|S46534 ubiquinol-cytochrome-c red... 29 3.1
CN825587 29 3.1
BF177810 29 4.0
CB828722 29 4.0
TC16745 similar to UP|Q94A14 (Q94A14) At2g43170/F14B2.11, partia... 28 8.9
TC13441 similar to UP|Q9SEH8 (Q9SEH8) Mannosyl-oligosaccharide 1... 28 8.9
>TC16160
Length = 1164
Score = 168 bits (425), Expect = 4e-42
Identities = 116/301 (38%), Positives = 164/301 (53%), Gaps = 19/301 (6%)
Frame = +2
Query: 49 YRSGSAPPTVEGSLSAFGSLRNFDYRANNSGR------SNNNDGVLTEDEIRSHPAYLSY 102
YRSGSAPPTVEGSL+A G L +G +N+ + +E+E+RS PAYL Y
Sbjct: 281 YRSGSAPPTVEGSLNAVGGLFGGGGGGGLAGPFSEFPGANHENDFASEEELRSDPAYLQY 460
Query: 103 YYSHESINPRLPPPLLSKEDWRVAQRFQAGGGSSSIERFGDWRK-NATSNGDSSSLFSMQ 161
YYS+ ++NPRLPPPLLSKEDWR AQR GG GD RK N + + SLF+
Sbjct: 461 YYSNVNLNPRLPPPLLSKEDWRSAQRASVLGG------IGDRRKVNRVDDSGARSLFATP 622
Query: 162 PGFSVQQAENDLMELRKASGRNLPRQSSTQLLDRHMDGMTRMPGTSLGVRRTCYSDILQD 221
PGF+ +++E++ A + +S + DG+ +PG LG ++ ++I QD
Sbjct: 623 PGFNSRKSESE------AENEKMRGSASAEW---GGDGLIGLPGLGLGSKQKSLAEIFQD 775
Query: 222 GF-DQPTLSSNMSRPASHNAF---VDIRDSTG------IVDREPLEGLRSSASTPGLVGL 271
+ T++ SRPAS NAF VDI S D + LRS + G
Sbjct: 776 DMGHKTTIAGFHSRPASRNAFDENVDIMTSAEADLADLCRDSSATDALRSGTNLQGSSAT 955
Query: 272 QNHGV-NSHNFSSVVGSSLSRSTTPE-SHVIGRPVGSGVPQMGSKVFSAENIGLGNHNGH 329
QN+G +S+ +++ +GS LSRSTTP+ HV P P G +V +AE G+GN + +
Sbjct: 956 QNNGTQSSYTYAAALGSPLSRSTTPDPQHVARAPSPCLTPIGGGRVAAAEKRGMGNPDAY 1135
Query: 330 S 330
+
Sbjct: 1136N 1138
>TC20139 similar to GB|AAG31805.1|11139702|AF315590 Pumilio 2 {Mus
musculus;} , partial (7%)
Length = 425
Score = 148 bits (373), Expect = 5e-36
Identities = 68/97 (70%), Positives = 83/97 (85%)
Frame = +2
Query: 844 CDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQ 903
C D TQ+ +M+EI+ +V LAQDQYGNYV+QH+LEHGKP+ER+ +I +L G IV+MSQQ
Sbjct: 98 CKDPNTQQKVMDEILGAVSMLAQDQYGNYVVQHVLEHGKPHERSAIIKELTGNIVQMSQQ 277
Query: 904 KFASNVIEKCLAFGSPEERQILVNEMLGTSDENEPLQ 940
KFASNV+EKCLAFG P ERQ LVNEMLG++DENEPLQ
Sbjct: 278 KFASNVVEKCLAFGGPAERQFLVNEMLGSTDENEPLQ 388
Score = 53.1 bits (126), Expect = 2e-07
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 1/83 (1%)
Frame = +2
Query: 685 YSSGFIQQKLETAS-VEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELAN 743
Y IQ+ LE + K+ EIL L D +GNYV+Q EHG +R +
Sbjct: 62 YGCRVIQRVLEHCKDPNTQQKVMDEILGAVSMLAQDQYGNYVVQHVLEHGKPHERSAIIK 241
Query: 744 QLTGHVLPLSLQMYGCRVIQKLL 766
+LTG+++ +S Q + V++K L
Sbjct: 242 ELTGNIVQMSQQKFASNVVEKCL 310
>AV409964
Length = 434
Score = 66.2 bits (160), Expect(2) = 1e-15
Identities = 31/44 (70%), Positives = 37/44 (83%)
Frame = +3
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQK 728
Y S FIQQKLETA+++EK +F EI+P A +LMTDVFGNYVIQK
Sbjct: 303 YGSRFIQQKLETATMDEKNMVFHEIMPQALSLMTDVFGNYVIQK 434
Score = 34.3 bits (77), Expect(2) = 1e-15
Identities = 16/26 (61%), Positives = 18/26 (68%)
Frame = +1
Query: 662 PCSMSSRTTKPNLLNFQISLIMWYSS 687
PCSMSS+ TKPN LN Q L M +S
Sbjct: 214 PCSMSSKATKPNALNLQKLLAMLLTS 291
Score = 32.0 bits (71), Expect = 0.47
Identities = 14/43 (32%), Positives = 23/43 (52%)
Frame = +3
Query: 833 YSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQ 875
Y S ++Q + + + ++ EIM +L D +GNYVIQ
Sbjct: 303 YGSRFIQQKLETATMDEKNMVFHEIMPQALSLMTDVFGNYVIQ 431
>AV770398
Length = 559
Score = 38.1 bits (87), Expect = 0.007
Identities = 19/28 (67%), Positives = 22/28 (77%)
Frame = -3
Query: 635 **EVLWEGLAAHGMLILAITWRVDLLHP 662
**EV E ++AHG+LILAI W DLLHP
Sbjct: 557 **EVHCEVMSAHGILILAINWTQDLLHP 474
>TC14364 weakly similar to UP|Q9SVZ7 (Q9SVZ7) Pumilio-like protein, partial
(8%)
Length = 702
Score = 35.8 bits (81), Expect = 0.033
Identities = 16/50 (32%), Positives = 27/50 (54%)
Frame = +3
Query: 716 LMTDVFGNYVIQKFFEHGTDSQRKELANQLTGHVLPLSLQMYGCRVIQKL 765
+M D F NYV+QK + +++QR L + + H L YG ++ +L
Sbjct: 54 MMKDPFANYVVQKVIDLCSENQRATLLSHIRVHAQALKKYTYGKHIVARL 203
Score = 30.4 bits (67), Expect = 1.4
Identities = 13/63 (20%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Frame = +3
Query: 851 EII-MEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNV 909
EII +E ++ T+ +D + NYV+Q +++ N+R ++S + + + + ++
Sbjct: 12 EIIGQDEQNNNLLTMMKDPFANYVVQKVIDLCSENQRATLLSHIRVHAQALKKYTYGKHI 191
Query: 910 IEK 912
+ +
Sbjct: 192 VAR 200
>TC8287 UP|AAQ87022 (AAQ87022) VDAC2.1, complete
Length = 1194
Score = 32.3 bits (72), Expect = 0.36
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 5/74 (6%)
Frame = +1
Query: 666 SSRTTKPNLLNFQISLIMWYSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNY- 724
S+ K L ++ + Y + I KL+TAS+ T F ++LP +A+ + +Y
Sbjct: 217 STAVKKGGLSVGDVAALYKYKNTLIDVKLDTASIISTTLTFTDLLPSTKAIASFKLPDYN 396
Query: 725 ----VIQKFFEHGT 734
+Q F +H T
Sbjct: 397 SGKLEVQYFHDHAT 438
>TC10692 weakly similar to UP|BAD08916 (BAD08916) Calcium-binding EF-hand
family protein-like, partial (32%)
Length = 634
Score = 31.6 bits (70), Expect = 0.62
Identities = 20/58 (34%), Positives = 30/58 (51%)
Frame = +2
Query: 83 NNDGVLTEDEIRSHPAYLSYYYSHESINPRLPPPLLSKEDWRVAQRFQAGGGSSSIER 140
NNDGVL+ E+R+ + +H I+ PP L+K V F G GS +++R
Sbjct: 131 NNDGVLSRAELRTAFESMRLIEAHFGIDVATPPEQLTKIYDSVFDTFD-GDGSGTVDR 301
>TC18291 similar to UP|TRMU_BORBU (O51625) Probable tRNA
(5-methylaminomethyl-2-thiouridylate)-methyltransferase
, partial (19%)
Length = 783
Score = 29.6 bits (65), Expect = 2.3
Identities = 17/61 (27%), Positives = 31/61 (49%), Gaps = 9/61 (14%)
Frame = +2
Query: 828 SCCTFYSSLWVQS-----YSDC---DDLKTQEIIMEEIMQSVCTL-AQDQYGNYVIQHIL 878
SC FY +W Q +S+C +DLK + + ++ + + D+Y N V+ +I+
Sbjct: 350 SCTAFYLKIWFQEDFENYWSECPWEEDLKFAKAVCNQVEVPLQVVHLTDEYWNNVVSYII 529
Query: 879 E 879
E
Sbjct: 530 E 532
>TC8273 UP|BAD11134 (BAD11134) Glutamate-rich protein, complete
Length = 923
Score = 29.3 bits (64), Expect = 3.1
Identities = 22/79 (27%), Positives = 38/79 (47%)
Frame = -1
Query: 276 VNSHNFSSVVGSSLSRSTTPESHVIGRPVGSGVPQMGSKVFSAENIGLGNHNGHSSNMTD 335
++S +FSS G++ + + S IG S GS VFSA + ++
Sbjct: 470 LSSDDFSSSTGAASVAAVSLVSSTIGSSFTSSGFTSGSSVFSASGL-----------VSS 324
Query: 336 LADMVSSLSGLNLSGARRA 354
+ +VS+ +G ++SGA A
Sbjct: 323 VTSLVSTSAGASVSGAAGA 267
>TC7906 homologue to PIR|S46534|S46534 ubiquinol-cytochrome-c reductase
Rieske iron-sulfur protein - potato
{Solanum tuberosum;} , partial (80%)
Length = 1179
Score = 29.3 bits (64), Expect = 3.1
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 1/62 (1%)
Frame = -3
Query: 237 SHNAFVDIRDSTGIVDREPLEGLRSSASTPGLVGLQNHGVNSHNFSSVV-GSSLSRSTTP 295
SH+ F+ + +S REP L ++ + ++ L G H +VV GS L RST+
Sbjct: 703 SHSGFLTLSESCWGSRREPTSTLFANLISSSVLRLMKTGFPRHLTVTVVPGSMLERSTSS 524
Query: 296 ES 297
E+
Sbjct: 523 EA 518
>CN825587
Length = 663
Score = 29.3 bits (64), Expect = 3.1
Identities = 10/23 (43%), Positives = 18/23 (77%)
Frame = -2
Query: 874 IQHILEHGKPNERTIVISKLAGQ 896
++H+LEHG+P++R ++ S L Q
Sbjct: 557 LKHLLEHGQPHDRLLISSSLLKQ 489
>BF177810
Length = 431
Score = 28.9 bits (63), Expect = 4.0
Identities = 22/80 (27%), Positives = 36/80 (44%), Gaps = 3/80 (3%)
Frame = +1
Query: 371 HANVMLSTPNNVNLPKHNELATDLNTFSLNERVNL---LKKTASYANLRSNAHSTGNLTS 427
+A V + PN NLP +N + + N + NL + S N+ +N ++ GN
Sbjct: 142 NAQVAFAKPNGANLPLNNGVPQNNNNAGILNNNNLPFITGLSGSTGNVFNNNNNNGN--G 315
Query: 428 IDFAGQVPSAYPANTTLNNV 447
F+ P+ PA TL +
Sbjct: 316 NGFSVTNPNNLPAGLTLQKL 375
>CB828722
Length = 529
Score = 28.9 bits (63), Expect = 4.0
Identities = 22/91 (24%), Positives = 40/91 (43%), Gaps = 22/91 (24%)
Frame = -1
Query: 812 CPSE*NSVYHHVFLWA-SCCTFYSSL-------WVQSYSDCDDLKTQEIIMEEIMQ---- 859
CPS+* S HHV + SCC S + + + +DLK + I+++ +
Sbjct: 382 CPSK*GSYRHHVVRTSRSCCIHVSQFCRVVTNPHIYLFGEANDLKMEANIIDDFRKGSDL 203
Query: 860 ----------SVCTLAQDQYGNYVIQHILEH 880
++ L Q Q+ N+++Q + H
Sbjct: 202 QLIM*MRTCINLLLLLQFQFFNFLLQSLRIH 110
>TC16745 similar to UP|Q94A14 (Q94A14) At2g43170/F14B2.11, partial (21%)
Length = 996
Score = 27.7 bits (60), Expect = 8.9
Identities = 12/30 (40%), Positives = 21/30 (70%), Gaps = 1/30 (3%)
Frame = -1
Query: 662 PCSMSSRTTKPNLLNFQIS-LIMWYSSGFI 690
PC ++ + + L+ FQI+ +I+WYS GF+
Sbjct: 429 PCRHAANSCQCPLIRFQITAIIIWYS*GFL 340
>TC13441 similar to UP|Q9SEH8 (Q9SEH8) Mannosyl-oligosaccharide
1,2-alpha-mannosidase , partial (10%)
Length = 576
Score = 27.7 bits (60), Expect = 8.9
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = +1
Query: 96 HPAYLSYYYSHESINPRLPPPLL 118
HP + S + ESINPR P LL
Sbjct: 112 HPQFNSLFTERESINPRTEPVLL 180
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.139 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,865,510
Number of Sequences: 28460
Number of extensions: 246220
Number of successful extensions: 1602
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1575
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1596
length of query: 949
length of database: 4,897,600
effective HSP length: 99
effective length of query: 850
effective length of database: 2,080,060
effective search space: 1768051000
effective search space used: 1768051000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144720.3


