

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.9 + phase: 0
(398 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
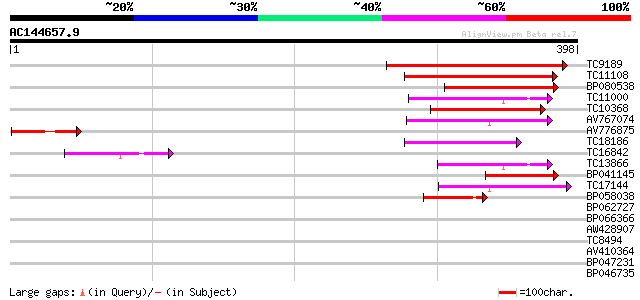
Score E
Sequences producing significant alignments: (bits) Value
TC9189 similar to UP|Q9FFN2 (Q9FFN2) Similarity to limonene cycl... 163 4e-41
TC11108 weakly similar to UP|O64805 (O64805) T1F15.13 protein (F... 115 1e-26
BP080538 83 9e-17
TC11000 homologue to UP|Q940Q8 (Q940Q8) AT5g61840/mac9_140, part... 71 3e-13
TC10368 71 4e-13
AV767074 65 3e-11
AV776875 64 6e-11
TC18186 similar to UP|Q8S9K6 (Q8S9K6) At1g21480/F24J8_23, partia... 62 2e-10
TC16842 59 2e-09
TC13866 homologue to UP|Q940Q8 (Q940Q8) AT5g61840/mac9_140, part... 50 7e-07
BP041145 49 2e-06
TC17144 42 1e-04
BP058038 40 7e-04
BP062727 33 0.064
BP066366 33 0.083
AW428907 32 0.14
TC8494 similar to UP|Q820I0 (Q820I0) Rnc; ribonuclease III (RNas... 31 0.32
AV410364 31 0.32
BP047231 30 0.92
BP046735 29 1.2
>TC9189 similar to UP|Q9FFN2 (Q9FFN2) Similarity to limonene cyclase,
partial (30%)
Length = 544
Score = 163 bits (413), Expect = 4e-41
Identities = 74/127 (58%), Positives = 95/127 (74%)
Frame = +3
Query: 265 LPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQF 324
LPK Y ++ SKFCLCPSG+EVASPRVVEAIY GCVPV+I ++Y PFSDVLNW F
Sbjct: 12 LPKAVSYYDMLRKSKFCLCPSGYEVASPRVVEAIYTGCVPVLISEHYVPPFSDVLNWKSF 191
Query: 325 SMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLR 384
S+E++V IP +K IL ++ +Y + V +R+HFE++ P K FD+ HM+LHSVWLR
Sbjct: 192 SLEVSVKDIPRLKEILLSVNTRQYIRMQRRVGHIRRHFEIHSPPKRFDVFHMVLHSVWLR 371
Query: 385 RLNFRLH 391
RLNFR+H
Sbjct: 372 RLNFRVH 392
>TC11108 weakly similar to UP|O64805 (O64805) T1F15.13 protein (Fragment),
partial (10%)
Length = 517
Score = 115 bits (288), Expect = 1e-26
Identities = 44/107 (41%), Positives = 78/107 (72%)
Frame = +1
Query: 278 SKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIK 337
SKFC+CP G +V S R+ ++I+ GC+PVI+ D Y LPF+D++NW +F++ + + ++K
Sbjct: 19 SKFCICPGGSQVNSARIADSIHYGCIPVILSDYYDLPFNDIINWRKFAVVLKESDVYQLK 198
Query: 338 TILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLR 384
IL+NI++ ++ L++N+ +V+KHF+ N P +D HM+++ +WLR
Sbjct: 199 QILKNISQAEFVTLHNNLVKVQKHFQWNSPPVGYDAFHMVMYDLWLR 339
>BP080538
Length = 387
Score = 82.8 bits (203), Expect = 9e-17
Identities = 37/80 (46%), Positives = 54/80 (67%)
Frame = -1
Query: 306 IICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRKHFEMN 365
II + Y LPF+DVLNW FS+ + IP +K IL+ I+ +Y +L SNV +VRKHF+ +
Sbjct: 387 IIANYYDLPFADVLNWKSFSVIVTTLDIPLLKKILKGISSDEYLMLQSNVLKVRKHFQWH 208
Query: 366 RPAKPFDLIHMILHSVWLRR 385
P FD +M+++ +WLRR
Sbjct: 207 SPPHDFDAFYMVMYELWLRR 148
>TC11000 homologue to UP|Q940Q8 (Q940Q8) AT5g61840/mac9_140, partial (33%)
Length = 637
Score = 71.2 bits (173), Expect = 3e-13
Identities = 38/105 (36%), Positives = 59/105 (56%), Gaps = 4/105 (3%)
Frame = +3
Query: 281 CLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTIL 340
CLCP G SPR+VEA+ GC+PVII D+ LPF+D + W + + + +P++ TIL
Sbjct: 3 CLCPLGWAPWSPRLVEAVVFGCIPVIIADDIVLPFADAIPWEEIGVFVDEKDVPQLDTIL 182
Query: 341 QNITE----TKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSV 381
+I K R+L + + F +PA+P D H +L+ +
Sbjct: 183 TSIPPEVILRKQRLLANPSMKQAMLFP--QPAQPGDTFHQVLNGL 311
>TC10368
Length = 561
Score = 70.9 bits (172), Expect = 4e-13
Identities = 34/81 (41%), Positives = 50/81 (60%)
Frame = +2
Query: 296 EAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNV 355
EAI CVPVII DN+ PF +VLNW F++ + IP +K IL +I E +Y L V
Sbjct: 2 EAISYECVPVIISDNFVPPFFEVLNWESFAVIVLEKDIPNLKNILLSIPEPRYLRLLMRV 181
Query: 356 RRVRKHFEMNRPAKPFDLIHM 376
++V++HF ++ +D+ HM
Sbjct: 182 KKVKQHFLWHKNPVQYDIFHM 244
>AV767074
Length = 481
Score = 64.7 bits (156), Expect = 3e-11
Identities = 33/106 (31%), Positives = 58/106 (54%), Gaps = 3/106 (2%)
Frame = -1
Query: 279 KFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPE--- 335
KFCL P+G +S R+ +AI + CVPVI+ D LPF D +++SQFS+ + +
Sbjct: 481 KFCLHPAGDTPSSCRLFDAIVSHCVPVIVSDQIELPFEDEIDYSQFSLFFSFKEALQPGY 302
Query: 336 IKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSV 381
+ L+ + K+ ++ ++ + H+E P K D ++M+ V
Sbjct: 301 MIDQLRKFPKDKWSEMWRQLKNISHHYEFQYPPKKEDAVNMLWRQV 164
>AV776875
Length = 611
Score = 63.5 bits (153), Expect = 6e-11
Identities = 34/49 (69%), Positives = 40/49 (81%)
Frame = +2
Query: 2 TSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQ 50
TSLE+IEE LA++RA+IQ+AIRSK K+SFV KGSIYLNP AFHQ
Sbjct: 299 TSLERIEECLARSRAMIQKAIRSK------KKSFVSKGSIYLNPQAFHQ 427
>TC18186 similar to UP|Q8S9K6 (Q8S9K6) At1g21480/F24J8_23, partial (32%)
Length = 606
Score = 61.6 bits (148), Expect = 2e-10
Identities = 32/83 (38%), Positives = 49/83 (58%), Gaps = 1/83 (1%)
Frame = +3
Query: 278 SKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRI-PEI 336
SKFCL P G + R E+ + CVPVII D LPF +V+++SQ S++ +RI PE+
Sbjct: 12 SKFCLAPRGESSWTLRFYESFFVECVPVIISDQIELPFQNVIDYSQISIKWPSNRIGPEL 191
Query: 337 KTILQNITETKYRVLYSNVRRVR 359
L++I + + + R+VR
Sbjct: 192 LQYLESIPDQNIEAIIARGRKVR 260
>TC16842
Length = 563
Score = 58.5 bits (140), Expect = 2e-09
Identities = 28/79 (35%), Positives = 45/79 (56%), Gaps = 2/79 (2%)
Frame = +3
Query: 39 GSIYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGP--VNNKYSIEGQFIDEMDTSNK 96
G +Y +P F ++ EMVK FKV++Y +G+ + P + KY+ EG F + +
Sbjct: 336 GDVYHSPRVFKLNYAEMVKNFKVYIYPDGDPKTFYQTPRKLTGKYASEGYFFQNI---RE 506
Query: 97 SPFKATHPELAHVFFLPFS 115
S F+ P+ AH+FF+P S
Sbjct: 507 SRFRTLDPDQAHLFFIPIS 563
>TC13866 homologue to UP|Q940Q8 (Q940Q8) AT5g61840/mac9_140, partial (28%)
Length = 542
Score = 50.1 bits (118), Expect = 7e-07
Identities = 27/85 (31%), Positives = 46/85 (53%), Gaps = 4/85 (4%)
Frame = +2
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITE----TKYRVLYSNVR 356
GC+PVII D+ LPF+D + W + + + +P++ TIL +I K R+L +
Sbjct: 2 GCIPVIIADDIVLPFADAIPWEDIGVFVDEEDVPKLDTILTSIPPEIILRKQRLLANPSM 181
Query: 357 RVRKHFEMNRPAKPFDLIHMILHSV 381
+ F +PA+P D H +L+ +
Sbjct: 182 KQAMLFP--QPAQPGDAFHQVLNGL 250
>BP041145
Length = 507
Score = 48.5 bits (114), Expect = 2e-06
Identities = 20/51 (39%), Positives = 33/51 (64%)
Frame = -3
Query: 335 EIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRR 385
++K IL++I E ++ L N+ +V+KHF N P D HM+++ +WLRR
Sbjct: 487 KLKDILRSIPEKQFVALNQNLVKVQKHFNWNTPPVRLDAFHMVMYELWLRR 335
>TC17144
Length = 602
Score = 42.4 bits (98), Expect = 1e-04
Identities = 20/96 (20%), Positives = 51/96 (52%), Gaps = 3/96 (3%)
Frame = +3
Query: 302 CVPVIICDNYSLPFSDVLNWSQFSMEI-AVDRIPE--IKTILQNITETKYRVLYSNVRRV 358
C+P I+ D LPF +L++ + ++ + ++D + + L+++ + + L N+ +
Sbjct: 39 CIPAIVSDELELPFEGILDYRKIALFVSSIDALKPGWLLKFLKDVRPAQIKELQPNLAQY 218
Query: 359 RKHFEMNRPAKPFDLIHMILHSVWLRRLNFRLHLKQ 394
+HF + PA+P ++ + + +N +LH ++
Sbjct: 219 SRHFVFSNPAQPLGPEDLVWKMMAGKVVNIKLHSRR 326
>BP058038
Length = 552
Score = 40.0 bits (92), Expect = 7e-04
Identities = 16/45 (35%), Positives = 30/45 (66%)
Frame = +3
Query: 291 SPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPE 335
+PR+VEA+ G +P+I+ ++ LPF+D + W + + VD+ P+
Sbjct: 279 NPRLVEAVVYGRIPIIVAEDIVLPFADAIPWVEIG--VFVDQTPQ 407
>BP062727
Length = 478
Score = 33.5 bits (75), Expect = 0.064
Identities = 11/34 (32%), Positives = 22/34 (64%)
Frame = +2
Query: 302 CVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPE 335
C+P+++ D+ LPF+D + W + + VD+ P+
Sbjct: 308 CIPIVVADDIVLPFADAIPWEEIC--VFVDQTPQ 403
>BP066366
Length = 448
Score = 33.1 bits (74), Expect = 0.083
Identities = 17/57 (29%), Positives = 26/57 (44%)
Frame = +2
Query: 271 YTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSME 327
Y +L G + +C + E + +A+ C P ICD Y S+V NW S +
Sbjct: 149 YCRLRGYNAIYICGTD-EYGTATETKAMEENCSPKEICDKYHAIHSEVYNWFNISFD 316
>AW428907
Length = 329
Score = 32.3 bits (72), Expect = 0.14
Identities = 11/28 (39%), Positives = 20/28 (71%)
Frame = +3
Query: 296 EAIYAGCVPVIICDNYSLPFSDVLNWSQ 323
+++ GC+PV+I D LP+ +VLN+ +
Sbjct: 237 DSVLQGCIPVVIQDGIFLPYENVLNYDR 320
>TC8494 similar to UP|Q820I0 (Q820I0) Rnc; ribonuclease III (RNase III)
protein , partial (7%)
Length = 597
Score = 31.2 bits (69), Expect = 0.32
Identities = 26/89 (29%), Positives = 42/89 (46%), Gaps = 2/89 (2%)
Frame = +1
Query: 52 HKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSI-EGQFIDEMDTSNKSPFKATHPELAHVF 110
H E+ K WVY+E L+H N S+ EG + +N+SP + H + A F
Sbjct: 31 HVEVKKCGHKWVYEEDLDQLIHSMEHNGNSSVLEGNLL-----TNESPDELNHGKAA*QF 195
Query: 111 -FLPFSVSKVIRYVYKPRKSRSDYNPHRL 138
F SV +++ + K +S+ + RL
Sbjct: 196 HFRYISVKQILIHFSKLSRSQGEKVQTRL 282
>AV410364
Length = 370
Score = 31.2 bits (69), Expect = 0.32
Identities = 17/51 (33%), Positives = 22/51 (42%), Gaps = 3/51 (5%)
Frame = +2
Query: 126 PRKSRSDYNPHRLQLLVEDYIKI---VANKYPYWNISQGADHFLLSCHDWG 173
P S ++ R L +E Y + +YPYWN S G DH D G
Sbjct: 50 PHLSMEEHKGLRSSLTLEYYKNAYHHIVEQYPYWNRSSGRDHIWFFSWDEG 202
>BP047231
Length = 476
Score = 29.6 bits (65), Expect = 0.92
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = -1
Query: 273 KLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPV 305
K+ S FCL P G + +++ AGCVPV
Sbjct: 212 KVFEKSVFCLQPPGDSYTRRSIFDSMLAGCVPV 114
>BP046735
Length = 535
Score = 29.3 bits (64), Expect = 1.2
Identities = 20/84 (23%), Positives = 36/84 (42%), Gaps = 6/84 (7%)
Frame = -2
Query: 278 SKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYS-LPFSDVLNWSQFSMEIAVDRI--- 333
S+FCL P G + + + AG +PV + LP+ L S + +DR
Sbjct: 525 SEFCLQPRGDSFTRRSIFDCMVAGSIPVFFWRRTAYLPYPWFLPGEPGSYSVYIDRNAVK 346
Query: 334 --PEIKTILQNITETKYRVLYSNV 355
+K +L+ ++ + R + V
Sbjct: 345 NGTSVKAVLEGFSQEEVRRMREKV 274
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,821,437
Number of Sequences: 28460
Number of extensions: 116786
Number of successful extensions: 602
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 597
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 598
length of query: 398
length of database: 4,897,600
effective HSP length: 92
effective length of query: 306
effective length of database: 2,279,280
effective search space: 697459680
effective search space used: 697459680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144657.9


