

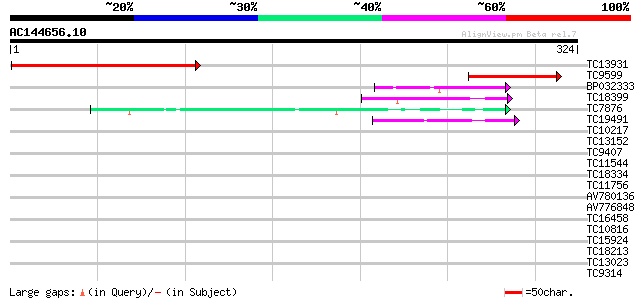
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.10 - phase: 0
(324 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13931 weakly similar to UP|BAC25162 (BAC25162) Adult male test... 172 7e-44
TC9599 92 9e-20
BP032333 43 8e-05
TC18399 42 2e-04
TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-bi... 40 5e-04
TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Ar... 40 7e-04
TC10217 homologue to UP|AAQ54906 (AAQ54906) Cell cycle switch pr... 39 0.001
TC13152 similar to UP|Q84XX6 (Q84XX6) Leunig (Fragment), partial... 37 0.005
TC9407 weakly similar to UP|TAF5_YEAST (P38129) Transcription in... 35 0.017
TC11544 homologue to UP|Q9SW94 (Q9SW94) G protein beta subunit, ... 34 0.038
TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1 (AT3... 33 0.050
TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snR... 33 0.050
AV780136 33 0.065
AV776848 33 0.085
TC16458 similar to UP|O48679 (O48679) F3I6.5 protein, partial (20%) 32 0.11
TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subun... 32 0.11
TC15924 weakly similar to GB|BAC21577.1|24060129|AP005438 G-prot... 32 0.11
TC18213 weakly similar to UP|Q9FIV2 (Q9FIV2) Similarity to pre-m... 30 0.42
TC13023 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragm... 30 0.42
TC9314 similar to GB|AAO63283.1|28950719|BT005219 At1g80650 {Ara... 29 1.2
>TC13931 weakly similar to UP|BAC25162 (BAC25162) Adult male testis cDNA,
RIKEN full-length enriched library, clone:1700094L18
product:protamine 1, full insert sequence, partial (47%)
Length = 392
Score = 172 bits (436), Expect = 7e-44
Identities = 82/108 (75%), Positives = 98/108 (89%)
Frame = +3
Query: 2 SSKRRISLVAGSYERFIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDT 61
+++RRI+LVAGSYERFIWGFNLNPN QTL P+FSYPSHLS+IKT+AV SVVASGG+DDT
Sbjct: 69 TTRRRINLVAGSYERFIWGFNLNPNRQTLKPIFSYPSHLSVIKTLAVCGSVVASGGTDDT 248
Query: 62 IHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSL 109
IHLY+LS +SSLGSL DHS+TVTAL+F+S PN+P P L+SADADGS+
Sbjct: 249 IHLYNLSNASSLGSLHDHSATVTALAFHSLPNIPHPTTLISADADGSV 392
>TC9599
Length = 579
Score = 92.4 bits (228), Expect = 9e-20
Identities = 44/53 (83%), Positives = 48/53 (90%)
Frame = +3
Query: 263 ATGDDEPYLVASASSDGTIRAWDVRMAATEKSEPLAECKTQSRLTCLAGSSLK 315
AT D+PYLVASASSDGTIR WDVRMAATEK PLAECKT+SRLTCL+GSS+K
Sbjct: 6 ATEGDDPYLVASASSDGTIRVWDVRMAATEKPTPLAECKTESRLTCLSGSSMK 164
>BP032333
Length = 506
Score = 42.7 bits (99), Expect = 8e-05
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 3/81 (3%)
Frame = -2
Query: 209 KRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYC---IEEAHAARVKGIVVLSDEATG 265
+ +LC A +N L+ +G ED+ I W + + YC + E H VK + + D
Sbjct: 496 RSILCLAVVEN-LICSGSEDKTIRIWRGEGNE--YCCLGVLEGHRGPVKSLSAVVDRCDS 326
Query: 266 DDEPYLVASASSDGTIRAWDV 286
+ YLV S S D I+ W +
Sbjct: 325 SEASYLVYSGSLDCDIKVWQI 263
>TC18399
Length = 542
Score = 41.6 bits (96), Expect = 2e-04
Identities = 27/88 (30%), Positives = 43/88 (48%), Gaps = 2/88 (2%)
Frame = +1
Query: 202 LMELQCPKRVLCAAPAKNG--LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVL 259
L+E+ AA NG +L TG D +I A D+++G I+ AH A + ++ L
Sbjct: 283 LLEVHAHTESCRAARFINGGRVLLTGSPDCSILATDVETGSTITRIDNAHEAAINRLINL 462
Query: 260 SDEATGDDEPYLVASASSDGTIRAWDVR 287
++ VAS +G I+ WD+R
Sbjct: 463 TEST--------VASGDDEGCIKVWDIR 522
>TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1379
Score = 40.0 bits (92), Expect = 5e-04
Identities = 53/244 (21%), Positives = 96/244 (38%), Gaps = 4/244 (1%)
Frame = +3
Query: 47 AVSNSVVASGGSDDTIHLYDL--STSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSAD 104
++ N + S D TI L++ ++ HS V+ + F SP L +VSA
Sbjct: 411 SIDNRQIVSASRDRTIKLWNTLGECKYTIQDNDAHSDWVSCVRF-SPSTLQ--PTIVSAS 581
Query: 105 ADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFC 164
D ++ +++ TL+ H VN +A+ P G + + +D + +L G+R +
Sbjct: 582 WDRTVKVWNLTNCKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLY- 758
Query: 165 CRLDKEASIVRFDVSGDSFFM--AVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLL 222
LD + I S + +++ A + + + E ++ +L+ + A
Sbjct: 759 -SLDAGSIIHALCFSPNRYWLCAATETSIKIWDLESKSIVEDLKVDLKTEADAT------ 917
Query: 223 YTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIR 282
TGG + KV YC L+ A G + S +DG IR
Sbjct: 918 -TGG--------NTNKKKVIYCTS------------LNWSADGS----TLFSGYTDGVIR 1022
Query: 283 AWDV 286
W +
Sbjct: 1023VWAI 1034
>TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;}, partial (39%)
Length = 527
Score = 39.7 bits (91), Expect = 7e-04
Identities = 27/84 (32%), Positives = 40/84 (47%)
Frame = +1
Query: 208 PKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDD 267
P R L +P LL+T +D N+ +D + GK HA+ V + V D
Sbjct: 22 PVRSLVYSPYDPRLLFTASDDGNVHMYDAE-GKALVGTMSGHASWVLCVDVSPDGGA--- 189
Query: 268 EPYLVASASSDGTIRAWDVRMAAT 291
+A+ SSD T+R WD+ M A+
Sbjct: 190 ----IATGSSDRTVRLWDLAMRAS 249
Score = 33.9 bits (76), Expect = 0.038
Identities = 25/73 (34%), Positives = 37/73 (50%)
Frame = +1
Query: 76 LTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAI 135
L H V +L YSP + PR L +A DG++ ++DA+G + T+S H V + +
Sbjct: 4 LEGHFMPVRSL-VYSPYD---PRLLFTASDDGNVHMYDAEGKALVGTMSGHASWVLCVDV 171
Query: 136 HPSGKIALTVSRD 148
P G T S D
Sbjct: 172 SPDGGAIATGSSD 210
Score = 31.6 bits (70), Expect = 0.19
Identities = 18/85 (21%), Positives = 39/85 (45%)
Frame = +1
Query: 55 SGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDA 114
+ D +H+YD + +G+++ H+S V + P + + +D ++ ++D
Sbjct: 70 TASDDGNVHMYDAEGKALVGTMSGHASWVLCVDV-----SPDGGAIATGSSDRTVRLWDL 234
Query: 115 DGFVHLKTLSVHKKGVNDLAIHPSG 139
++T+S H V +A P G
Sbjct: 235 AMRASVQTMSNHTDQVWGVAFRPPG 309
>TC10217 homologue to UP|AAQ54906 (AAQ54906) Cell cycle switch protein
CCS52a, partial (45%)
Length = 640
Score = 38.9 bits (89), Expect = 0.001
Identities = 41/147 (27%), Positives = 64/147 (42%), Gaps = 7/147 (4%)
Frame = +2
Query: 17 FIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVS---NSVVASGG--SDDTIHLYDLSTSS 71
F+W N + PV Y H + +K +A S + ++ASGG +D I ++ +T+S
Sbjct: 191 FVW------NQHSTQPVLKYCEHTAAVKAIAWSPHLHGLLASGGGTADRCIRFWNTTTNS 352
Query: 72 SLGSLTDHSSTVTALSFYSPPNLPFPRNLVSAD--ADGSLAIFDADGFVHLKTLSVHKKG 129
L S D S V L + N LVS + + ++ L TL+ H
Sbjct: 353 HL-SCMDTGSQVCNLVWSKNVN-----ELVSTHGYSQNQIIVWRYPTMSKLATLTGHTYR 514
Query: 130 VNDLAIHPSGKIALTVSRDSCFAMVNL 156
V LAI P G+ +T + D N+
Sbjct: 515 VLYLAISPDGQTIVTGAGDETLXFWNV 595
>TC13152 similar to UP|Q84XX6 (Q84XX6) Leunig (Fragment), partial (60%)
Length = 453
Score = 37.0 bits (84), Expect = 0.005
Identities = 29/99 (29%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Frame = +2
Query: 52 VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAI 111
++ASGG D L+ + +L +HS+ +T + F P++P L ++ D ++ +
Sbjct: 2 LLASGGHDKKAVLWYTDSLKQKATLEEHSALITDVRF--SPSMP---RLATSSFDKTVKV 166
Query: 112 FDADG-FVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDS 149
+D D L+T + H V L HP+ K L S DS
Sbjct: 167 WDVDNPGYSLRTFTGHSASVMSLDFHPN-KEDLICSCDS 280
Score = 27.3 bits (59), Expect = 3.6
Identities = 18/65 (27%), Positives = 27/65 (40%)
Frame = +2
Query: 222 LYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTI 281
L T D+ + WD+ + + H+A V L +D L+ S SDG I
Sbjct: 131 LATSSFDKTVKVWDVDNPGYSLRTFTGHSASVMS---LDFHPNKED---LICSCDSDGEI 292
Query: 282 RAWDV 286
R W +
Sbjct: 293 RYWSI 307
>TC9407 weakly similar to UP|TAF5_YEAST (P38129) Transcription initiation
factor TFIID subunit 5 (TBP-associated factor 5)
(TBP-associated factor 90 kDa) (TAFII-90), partial (4%)
Length = 530
Score = 35.0 bits (79), Expect = 0.017
Identities = 20/62 (32%), Positives = 32/62 (51%)
Frame = +2
Query: 98 RNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLV 157
R L +A AD ++ I++ DGF KTL H++ V D G +T S D+ + ++
Sbjct: 53 RYLATASADHTVKIWNVDGFTLDKTLIGHQRWVWDCVFSVDGAYLITASSDTTARLWSMS 232
Query: 158 RG 159
G
Sbjct: 233 TG 238
>TC11544 homologue to UP|Q9SW94 (Q9SW94) G protein beta subunit, partial
(50%)
Length = 774
Score = 33.9 bits (76), Expect = 0.038
Identities = 22/79 (27%), Positives = 33/79 (40%), Gaps = 4/79 (5%)
Frame = +2
Query: 216 PAKNGLLYTGGEDRNITAWDLKSGKVAYCI----EEAHAARVKGIVVLSDEATGDDEPYL 271
P ++ L TG D+ WD+ +G + H A V I + + +
Sbjct: 554 PDEDTHLITGSGDQTCVLWDITTGLRTSVFGGEFQSGHTADVLSISINGSNSR------M 715
Query: 272 VASASSDGTIRAWDVRMAA 290
S S DGT R WD R+A+
Sbjct: 716 FVSGSCDGTARLWDTRVAS 772
>TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1
(AT3g18860/MCB22_3), partial (21%)
Length = 595
Score = 33.5 bits (75), Expect = 0.050
Identities = 26/89 (29%), Positives = 42/89 (46%)
Frame = +2
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
VASGG D + ++DLST +L H VT+++F ++VSA D +L +
Sbjct: 338 VASGGMDTLVLVWDLSTGEKAHTLKGHQLQVTSIAFDD-------GDVVSASVDCTLRRW 496
Query: 113 DADGFVHLKTLSVHKKGVNDLAIHPSGKI 141
+T HK V + P+G++
Sbjct: 497 KNGQCT--ETWEAHKAPVQAVIKLPTGEL 577
Score = 31.2 bits (69), Expect = 0.25
Identities = 22/65 (33%), Positives = 32/65 (48%)
Frame = +2
Query: 220 GLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDG 279
G + +GG D + WDL +G+ A+ + + H +V I GD V SAS D
Sbjct: 332 GGVASGGMDTLVLVWDLSTGEKAHTL-KGHQLQVTSIAF----DDGD-----VVSASVDC 481
Query: 280 TIRAW 284
T+R W
Sbjct: 482 TLRRW 496
>TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snRNP-specific
40 kDa protein (hPrp8-binding) {Homo sapiens;} , partial
(43%)
Length = 719
Score = 33.5 bits (75), Expect = 0.050
Identities = 20/75 (26%), Positives = 34/75 (44%)
Frame = +1
Query: 222 LYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTI 281
+ + D+ + WD+++GK + E H + V P LV S S DGT
Sbjct: 478 IVSASPDKTVRVWDVETGKQVKKMVE-HLSYVNSC------CPSRRGPPLVVSGSDDGTA 636
Query: 282 RAWDVRMAATEKSEP 296
+ WD+R + ++ P
Sbjct: 637 KLWDMRQRGSIQTFP 681
>AV780136
Length = 558
Score = 33.1 bits (74), Expect = 0.065
Identities = 19/66 (28%), Positives = 31/66 (46%)
Frame = -1
Query: 221 LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGT 280
+L +GG ++ I WD +SG + HA ++ +++ D S SSD
Sbjct: 531 VLVSGGTEKVIRVWDPRSGSKTLKL-RGHADNIRALLL-------DSTGRYCLSGSSDSM 376
Query: 281 IRAWDV 286
IR WD+
Sbjct: 375 IRLWDI 358
>AV776848
Length = 584
Score = 32.7 bits (73), Expect = 0.085
Identities = 28/115 (24%), Positives = 45/115 (38%), Gaps = 1/115 (0%)
Frame = +3
Query: 205 LQCPKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIE-EAHAARVKGIVVLSDEA 263
+ C + AA + L+TG D + W L V E+H V V++ D
Sbjct: 228 INCLALLTSAASDGSDYLFTGSRDGRLKRWALAEDAVTCSATFESHVDWVNDAVLVGDNT 407
Query: 264 TGDDEPYLVASASSDGTIRAWDVRMAATEKSEPLAECKTQSRLTCLAGSSLKCKL 318
+ S SSD T++ W+ A + S + +TCLA + C +
Sbjct: 408 --------LVSCSSDTTLKTWN---AFSVGSCTRTLRQHSDYVTCLAAAGKNCNI 539
>TC16458 similar to UP|O48679 (O48679) F3I6.5 protein, partial (20%)
Length = 539
Score = 32.3 bits (72), Expect = 0.11
Identities = 28/100 (28%), Positives = 40/100 (40%), Gaps = 8/100 (8%)
Frame = +3
Query: 193 VHQAEDARLLMELQCPKRVLCAAPAKN---GLLYTGGEDRNITAWDLKSG-----KVAYC 244
VH E L+E + A A N +LY+G DR+I W+ G V
Sbjct: 6 VHAGEKKHSLLETLEKHKSAVNALALNEDGSVLYSGACDRSILVWERDGGDGGGEMVVVG 185
Query: 245 IEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAW 284
H + +VV+ D LV S S+D ++R W
Sbjct: 186 ALRGHTKAILCLVVVGD---------LVFSGSADNSVRVW 278
>TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subunit-like
protein, partial (20%)
Length = 979
Score = 32.3 bits (72), Expect = 0.11
Identities = 41/193 (21%), Positives = 73/193 (37%), Gaps = 1/193 (0%)
Frame = +1
Query: 49 SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGS 108
S + SGG D I +++ L +L H + + F+ P+ +VSA D +
Sbjct: 424 SQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHES--PW---IVSASDDQT 588
Query: 109 LAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLD 168
+ I++ + L+ H V HP + ++ S D + ++ +R D
Sbjct: 589 IRIWNWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSLKRKNASPAD 768
Query: 169 KEASIVRFDVSGDSFFMAVDEIVN-VHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGGE 227
I+R F VD +V V + D + P A P L+ + +
Sbjct: 769 ---DILRLSQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHP-----ALP----LIVSAAD 912
Query: 228 DRNITAWDLKSGK 240
DR + W + K
Sbjct: 913 DRQVKLWRMNDTK 951
Score = 26.9 bits (58), Expect = 4.7
Identities = 11/26 (42%), Positives = 15/26 (57%)
Frame = +1
Query: 271 LVASASSDGTIRAWDVRMAATEKSEP 296
LV SAS D T+R WD+ + + P
Sbjct: 685 LVVSASLDQTVRVWDIGSLKRKNASP 762
Score = 26.9 bits (58), Expect = 4.7
Identities = 18/65 (27%), Positives = 31/65 (47%)
Frame = +1
Query: 221 LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGT 280
L +GG+D I W+ K + + + H ++ + + P++V SAS D T
Sbjct: 433 LFVSGGDDYKIKVWNYKLHRCLFTLL-GHLDYIRTVQF------HHESPWIV-SASDDQT 588
Query: 281 IRAWD 285
IR W+
Sbjct: 589 IRIWN 603
>TC15924 weakly similar to GB|BAC21577.1|24060129|AP005438 G-protein beta
family-like protein {Oryza sativa (japonica
cultivar-group);}, partial (11%)
Length = 1262
Score = 32.3 bits (72), Expect = 0.11
Identities = 24/89 (26%), Positives = 40/89 (43%), Gaps = 2/89 (2%)
Frame = +1
Query: 212 LCAAPAKNGLLYTGGEDRNITAWDLK--SGKVAYCIEEAHAARVKGIVVLSDEATGDDEP 269
+C+ GLL++G D +I WD++ S + + I+E K + S D
Sbjct: 88 VCSLIYYKGLLFSGYSDGSIKVWDIRGHSASLVWDIKE----HKKSVTCFSVHEPSDS-- 249
Query: 270 YLVASASSDGTIRAWDVRMAATEKSEPLA 298
+ S S+D TI+ W + E E +A
Sbjct: 250 --LLSGSTDKTIKVWKMIQRKLECVEVIA 330
>TC18213 weakly similar to UP|Q9FIV2 (Q9FIV2) Similarity to pre-mRNA
splicing factor, partial (25%)
Length = 662
Score = 30.4 bits (67), Expect = 0.42
Identities = 16/61 (26%), Positives = 29/61 (47%)
Frame = +1
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
+ASG SD +++LYD +S + + +SF+ P + S DG + +F
Sbjct: 136 LASGSSDGSVYLYDYHSSKVVKKIKAFDQACIDVSFHP----VIPNVIASCSWDGGILVF 303
Query: 113 D 113
+
Sbjct: 304 E 306
>TC13023 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragment),
partial (18%)
Length = 591
Score = 30.4 bits (67), Expect = 0.42
Identities = 15/35 (42%), Positives = 19/35 (53%), Gaps = 1/35 (2%)
Frame = +1
Query: 211 VLC-AAPAKNGLLYTGGEDRNITAWDLKSGKVAYC 244
V+C + GLL TGG DR + WD+ G YC
Sbjct: 418 VMCMSCHPSGGLLATGGADRKVLVWDVDGG---YC 513
>TC9314 similar to GB|AAO63283.1|28950719|BT005219 At1g80650 {Arabidopsis
thaliana;}, partial (11%)
Length = 579
Score = 28.9 bits (63), Expect = 1.2
Identities = 18/40 (45%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Frame = -3
Query: 142 ALTVSRDSCFAMVNL--VRGRRSFCCRLDKEASIVRFDVS 179
AL V DSC A L V GR FCCR + S F S
Sbjct: 223 ALVVVADSCSAASQLPVVLGR*PFCCRYQRAPSAALFAAS 104
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,623,009
Number of Sequences: 28460
Number of extensions: 73633
Number of successful extensions: 514
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 482
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 509
length of query: 324
length of database: 4,897,600
effective HSP length: 90
effective length of query: 234
effective length of database: 2,336,200
effective search space: 546670800
effective search space used: 546670800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC144656.10


