

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.8 - phase: 0
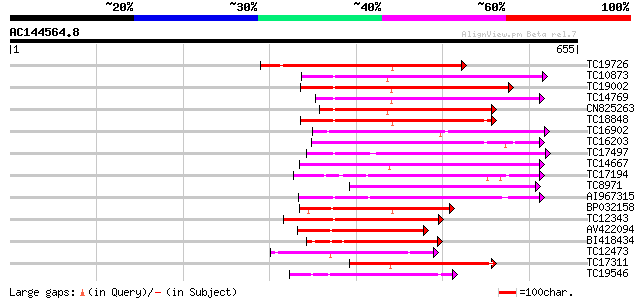
(655 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 214 5e-56
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 202 1e-52
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 200 5e-52
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 186 1e-47
CN825263 181 3e-46
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 176 1e-44
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 175 2e-44
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 171 3e-43
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 167 4e-42
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 166 1e-41
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 160 6e-40
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 157 5e-39
AI967315 157 7e-39
BP032158 153 1e-37
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 151 3e-37
AV422094 145 3e-35
BI418434 140 5e-34
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 139 1e-33
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 139 2e-33
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 133 1e-31
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 214 bits (544), Expect = 5e-56
Identities = 115/245 (46%), Positives = 159/245 (63%), Gaps = 7/245 (2%)
Frame = +3
Query: 290 LSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFE 349
L + +VA++ LW K+ E E ++ Q + + P++ +Y EL ATNNF+
Sbjct: 3 LGFVIVAVIGGLLWWIRLKKKRE---EGSNSQILGTLKSLPGTPREFNYVELKKATNNFD 173
Query: 350 ETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLT 409
E KLGQGG+G VY+G A+K S D + + +E+ II++LRH++LV+L
Sbjct: 174 EKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLSELIIINRLRHKHLVRLQ 353
Query: 410 GWCHKKNELILIYEYMPNGSLDFHLF--RGGSI---LPWNLRYNIALGLASALLYLQEEW 464
GWCHK L+L+Y+YMPNGSLD H+F GG+I L W LRY I G+ASAL YL E+
Sbjct: 354 GWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKIISGVASALNYLHNEY 533
Query: 465 EKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTV--VAGTRGYLAPEYIDT 522
++ V+HRD+K+SNIMLDS+FN KLGDFGLAR +++EK S T + V GT GY+APE T
Sbjct: 534 DQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEGVHGTMGYIAPECFHT 713
Query: 523 SKARK 527
KA +
Sbjct: 714 GKATR 728
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 202 bits (515), Expect = 1e-52
Identities = 119/289 (41%), Positives = 167/289 (57%), Gaps = 5/289 (1%)
Frame = +1
Query: 338 YYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKII 397
+ EL +AT NF+E +G+GGFG VYKG +VA +K++S D RQG +++ EV ++
Sbjct: 421 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVA-VKQLSHDGRQGFQEFVMEVLML 597
Query: 398 SQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIALGLA 454
S L H NLV+L G+C ++ +L+YEYMP GSL+ HLF L W+ R +A+G A
Sbjct: 598 SLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAA 777
Query: 455 SALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMD-HEKGSETTVVAGTRG 513
L YL + VI+RD+KS+NI+LD++FN KL DFGLA+L + +T V GT G
Sbjct: 778 RGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYG 957
Query: 514 YLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYG-LRNL 572
Y APEY + K +SDI+SFGVVLLE+ G++AI GE +LV W + R
Sbjct: 958 YCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRF 1137
Query: 573 IVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEA 621
DP L G F + L + + C RP I ++ L + A
Sbjct: 1138GHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA 1284
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 200 bits (509), Expect = 5e-52
Identities = 107/249 (42%), Positives = 154/249 (60%), Gaps = 3/249 (1%)
Frame = +1
Query: 337 SYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKI 396
S EL +ATNNF KLG+GGFG VY G D + +A +KR+ S + +++ EV+I
Sbjct: 31 SLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIA-VKRLKVWSNKADMEFAVEVEI 207
Query: 397 ISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGS---ILPWNLRYNIALGL 453
++++RH+NL+ L G+C + E +++Y+YMPN SL HL S +L WN R NIA+G
Sbjct: 208 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS 387
Query: 454 ASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRG 513
A ++YL + +IHRDIK+SN++LDSDF ++ DFG A+L+ TT V GT G
Sbjct: 388 AEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLG 567
Query: 514 YLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLI 573
YLAPEY KA + D+FSFG++LLE+A GKK + + S+ +W L +
Sbjct: 568 YLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKKFT 747
Query: 574 VAADPKLCG 582
ADP+L G
Sbjct: 748 EFADPRLNG 774
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 186 bits (472), Expect = 1e-47
Identities = 106/268 (39%), Positives = 156/268 (57%), Gaps = 4/268 (1%)
Frame = +3
Query: 354 LGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCH 413
+G+GGFG VY+G D VA +K S+ S QG +++ E+ ++S ++H NLV L G+C+
Sbjct: 2181 IGEGGFGSVYRGTLNDGQEVA-VKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCN 2357
Query: 414 KKNELILIYEYMPNGSLDFHLFRGGS---ILPWNLRYNIALGLASALLYLQEEWEKCVIH 470
+ ++ IL+Y +M NGSL L+ + IL W R +IALG A L YL + VIH
Sbjct: 2358 ESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIH 2537
Query: 471 RDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTV-VAGTRGYLAPEYIDTSKARKES 529
RDIKSSNI+LD K+ DFG ++ E S ++ V GT GYL PEY T + ++S
Sbjct: 2538 RDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKS 2717
Query: 530 DIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKLCGIFDVKQL 589
D+FSFGVVLLEI G++ ++ + E SLVEW + DP + G + + +
Sbjct: 2718 DVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAM 2897
Query: 590 ECLLVVGLWCANPDITSRPSIKKVIKVL 617
++ V L C P T RPS+ +++ L
Sbjct: 2898 WRVVEVALQCLEPFSTYRPSMVAIVREL 2981
>CN825263
Length = 663
Score = 181 bits (459), Expect = 3e-46
Identities = 100/209 (47%), Positives = 131/209 (61%), Gaps = 4/209 (1%)
Frame = +1
Query: 358 GFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNE 417
GFG VYKG D VA +K + D ++G +++ AEV+++S+L HRNLVKL G C +K
Sbjct: 1 GFGLVYKGILNDGRDVA-VKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 418 LILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIK 474
LIYE +PNGS++ HL + L WN R IALG A L YL E+ CVIHRD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 475 SSNIMLDSDFNTKLGDFGLAR-LMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFS 533
SSNI+L+ DF K+ DFGLAR +D +T V GT GYLAPEY T +SD++S
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 534 FGVVLLEIACGKKAIHHQELEGEVSLVEW 562
+GVVLLE+ G K + + G+ +LV W
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQENLVTW 624
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 176 bits (445), Expect = 1e-44
Identities = 94/231 (40%), Positives = 143/231 (61%), Gaps = 5/231 (2%)
Frame = +2
Query: 337 SYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKI 396
+Y EL AT F + KLG+GGFG VY G D +A +K++ A + + +++ EV++
Sbjct: 269 TYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIA-VKKLKAMNSKAEMEFAVEVEV 445
Query: 397 ISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI---LPWNLRYNIALGL 453
+ ++RH+NL+ L G+C ++ +++Y+YMPN SL HL ++ L W R IA+G
Sbjct: 446 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGS 625
Query: 454 ASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRG 513
A +LYL E +IHRDIK+SN++L+SDF + DFG A+L+ TT V GT G
Sbjct: 626 AEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTLG 805
Query: 514 YLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEV--SLVEW 562
YLAPEY K + D++SFG++LLE+ G+K I ++L G V ++ EW
Sbjct: 806 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPI--EKLPGGVKRTITEW 952
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 175 bits (444), Expect = 2e-44
Identities = 106/278 (38%), Positives = 154/278 (55%), Gaps = 4/278 (1%)
Frame = +3
Query: 350 ETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLT 409
E LG GGFG VY G D + AIKR + S QGV ++ E++++S+LRHR+LV L
Sbjct: 3 EALLLGVGGFGKVYYGEV-DGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLI 179
Query: 410 GWCHKKNELILIYEYMPNGSLDFHLFRGGSI-LPWNLRYNIALGLASALLYLQEEWEKCV 468
G+C + E+IL+Y++M G+L HL++ LPW R I +G A L YL + +
Sbjct: 180 GYCEENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTI 359
Query: 469 IHRDIKSSNIMLDSDFNTKLGDFGLARL---MDHEKGSETTVVAGTRGYLAPEYIDTSKA 525
IHRD+K++NI+LD + K+ DFGL++ +D+ S TVV G+ GYL PEY +
Sbjct: 360 IHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVS--TVVKGSFGYLDPEYFRRQQL 533
Query: 526 RKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKLCGIFD 585
+SD++SFGVVL EI C + A++ + +VSL EW Y L DP L G
Sbjct: 534 TDKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIA 713
Query: 586 VKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPL 623
+ + + C + RPS+ V+ L F L
Sbjct: 714 PECFKKFAETAMKCVSDQGIERPSMGDVLWNLEFALQL 827
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 171 bits (433), Expect = 3e-43
Identities = 97/279 (34%), Positives = 160/279 (56%), Gaps = 10/279 (3%)
Frame = +2
Query: 349 EETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKL 408
+E +G+GG G VY+G + VA + + S + + AE++ + ++RHRN+++L
Sbjct: 2195 KEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRL 2374
Query: 409 TGWCHKKNELILIYEYMPNGSLD--FHLFRGGSILPWNLRYNIALGLASALLYLQEEWEK 466
G+ K+ +L+YEYMPNGSL H +GG L W +RY IA+ A L Y+ +
Sbjct: 2375 LGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH-LRWEMRYKIAVEAARGLCYMHHDCSP 2551
Query: 467 CVIHRDIKSSNIMLDSDFNTKLGDFGLAR-LMDHEKGSETTVVAGTRGYLAPEYIDTSKA 525
+IHRD+KS+NI+LD+DF + DFGLA+ L D + +AG+ GY+APEY T K
Sbjct: 2552 LIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKV 2731
Query: 526 RKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRN-------LIVAADP 578
++SD++SFGVVLLE+ G+K + E V +V WV + + ++ DP
Sbjct: 2732 DEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDP 2905
Query: 579 KLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
+L G + + + + + + C +RP++++V+ +L
Sbjct: 2906 RLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 167 bits (424), Expect = 4e-42
Identities = 103/284 (36%), Positives = 157/284 (55%), Gaps = 2/284 (0%)
Frame = +1
Query: 343 NATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRH 402
+ATN+F + KLG+GGFG VYKG + +A +KR+S S QG++++ E+K+I++L+H
Sbjct: 1657 SATNHFSLSNKLGEGGFGPVYKGLLANGQEIA-VKRLSNTSGQGMEEFKNEIKLIARLQH 1833
Query: 403 RNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGS-ILPWNLRYNIALGLASALLYLQ 461
RNLVKL G ++E N + L S ++ WN R I G+A LLYL
Sbjct: 1834 RNLVKLFGCSVHQDE-----NSHANKKMKILLDSTRSKLVDWNKRLQIIDGIARGLLYLH 1998
Query: 462 EEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARL-MDHEKGSETTVVAGTRGYLAPEYI 520
++ +IHRD+K+SNI+LD + N K+ DFGLAR+ + + + T V GT GY+ PEY
Sbjct: 1999 QDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYA 2178
Query: 521 DTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKL 580
+SD+FSFGV++LEI GKK + ++L+ W L+ + D L
Sbjct: 2179 VHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEERPLELVDELL 2358
Query: 581 CGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLP 624
++ + V L C +RP + ++ +LN E LP
Sbjct: 2359 DDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGEKELP 2490
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 166 bits (419), Expect = 1e-41
Identities = 101/291 (34%), Positives = 152/291 (51%), Gaps = 9/291 (3%)
Frame = +2
Query: 336 ISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVK 395
+S EL T+NF +G+G +G VY D N+VA K + + ++ +V
Sbjct: 323 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVS 502
Query: 396 IISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLD--FHLFRG------GSILPWNLRY 447
++S+L++ N V+L G+C + N +L YE+ GSL H +G G L W R
Sbjct: 503 MVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRV 682
Query: 448 NIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLA-RLMDHEKGSETT 506
IA+ A L YL E+ + +IHRDI+SSN+++ D+ K+ DF L+ + D +T
Sbjct: 683 RIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHST 862
Query: 507 VVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWEL 566
V GT GY APEY T + ++SD++SFGVVLLE+ G+K + H G+ SLV W
Sbjct: 863 RVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPR 1042
Query: 567 YGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
+ DPKL G + K + L V C + RP++ V+K L
Sbjct: 1043LSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 160 bits (405), Expect = 6e-40
Identities = 103/299 (34%), Positives = 157/299 (52%), Gaps = 10/299 (3%)
Frame = +1
Query: 329 MGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVK 388
M A + SY EL ATNNF K+GQGGFG VY Y + AIK++ Q
Sbjct: 1030 MVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVY--YAELRGKKTAIKKMDV---QAST 1194
Query: 389 QYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI-LPWNLRY 447
++ E+K+++ + H NLV+L G+C + L L+YE++ NG+L +L G LPW+ R
Sbjct: 1195 EFLCELKVLTHVHHLNLVRLIGYC-VEGSLFLVYEHIDNGNLGQYLHGSGKEPLPWSSRV 1371
Query: 448 NIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTV 507
IAL A L Y+ E IHRD+KS+NI++D + K+ DFGL +L++ + T
Sbjct: 1372 QIALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTR 1551
Query: 508 VAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHH-----QELEGEVSLVEW 562
+ GT GY+ PEY + D+++FGVVL E+ K A+ E +G V+L E
Sbjct: 1552 LVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEE 1731
Query: 563 VWE----LYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
LR L+ DP+L + + + + +G C + RPS++ ++ L
Sbjct: 1732 ALNKSDPCDALRKLV---DPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVAL 1899
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 157 bits (397), Expect = 5e-39
Identities = 90/224 (40%), Positives = 134/224 (59%), Gaps = 3/224 (1%)
Frame = +3
Query: 393 EVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFR-GGSILPWNLRYNIAL 451
EV++++++ HRNLVKL G+ K NE ILI EY+PNG+L HL G IL +N R IA+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKILDFNQRLEIAI 185
Query: 452 GLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARL--MDHEKGSETTVVA 509
+A L YL EK +IHRD+KSSNI+L K+ DFG ARL ++ ++ +T V
Sbjct: 186 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTKVK 365
Query: 510 GTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGL 569
GT GYL PEY+ T + +SD++SFG++LLEI G++ + ++ E + W + Y
Sbjct: 366 GTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKYNE 545
Query: 570 RNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKV 613
+++ DP + + L +L + CA P T RP++K V
Sbjct: 546 GSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSV 677
>AI967315
Length = 1308
Score = 157 bits (396), Expect = 7e-39
Identities = 100/291 (34%), Positives = 154/291 (52%), Gaps = 7/291 (2%)
Frame = +1
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSR--QGVKQYS 391
K SY EL +ATN F +G+GG+ VYKG + + +A +KR++ R + K++
Sbjct: 106 KCFSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIA-VKRLTRTCRDERKEKEFL 282
Query: 392 AEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILP--WNLRYNI 449
E+ I + H N++ L G C N L L++E GS+ L + P W RY I
Sbjct: 283 TEIGTIGHVCHSNVMPLLGCCID-NGLYLVFELSTVGSVA-SLIHDEKMAPLDWKTRYKI 456
Query: 450 ALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTV-V 508
LG A L YL + ++ +IHRDIK+SNI+L DF ++ DFGLA+ + + + +
Sbjct: 457 VLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPI 636
Query: 509 AGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAI--HHQELEGEVSLVEWVWEL 566
GT G+LAPEY +++D+F+FGV LLE+ G+K + HQ L + WE+
Sbjct: 637 EGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKWEI 816
Query: 567 YGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
L DP+L G +DV Q + C T RP++ +V++V+
Sbjct: 817 EKL------VDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVM 951
>BP032158
Length = 555
Score = 153 bits (386), Expect = 1e-37
Identities = 82/185 (44%), Positives = 116/185 (62%), Gaps = 6/185 (3%)
Frame = +2
Query: 335 KISYYELLN---ATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYS 391
K Y+ L ATNNF+ K+G+GGFG VYKG + + V A+K++S+ S+QG +++
Sbjct: 2 KTGYFSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGD-VIAVKQLSSKSKQGNREFI 178
Query: 392 AEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI---LPWNLRYN 448
E+ +IS L+H NLVKL G C + N+L+L+YEYM N SL LF L W R
Sbjct: 179 NEIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMK 358
Query: 449 IALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVV 508
I +G+A L YL EE ++HRDIK++N++LD D K+ DFGLA+L + E +T +
Sbjct: 359 ICVGIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRI 538
Query: 509 AGTRG 513
AGT G
Sbjct: 539 AGTIG 553
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 151 bits (382), Expect = 3e-37
Identities = 80/189 (42%), Positives = 118/189 (62%), Gaps = 4/189 (2%)
Frame = +3
Query: 317 TTSDQDMDDEFQMGAGP--KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVA 374
++S+ +D+ Q A K SY L+ AT NF KLG+GGFG V+KG D +A
Sbjct: 144 SSSEAQNEDDIQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIA 323
Query: 375 AIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHL 434
+K++S S QG Q+ E K++++++HRN+V L G+C +E +L+YEY+P SLD L
Sbjct: 324 -VKKLSRRSNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLL 500
Query: 435 FRGGSI--LPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFG 492
FR L W R++I G+A LLYL E+ C+IHRDIK++NI+LD + K+ DFG
Sbjct: 501 FRSQKKEQLDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFG 680
Query: 493 LARLMDHEK 501
LAR+ ++
Sbjct: 681 LARIFPEDQ 707
>AV422094
Length = 462
Score = 145 bits (365), Expect = 3e-35
Identities = 75/151 (49%), Positives = 100/151 (65%)
Frame = +2
Query: 333 PKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSA 392
P SY EL NATN+F KLG+GGFG VYKG D +V A+K++S S QG Q+ A
Sbjct: 5 PYTFSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDG-TVIAVKQLSLGSHQGKSQFIA 181
Query: 393 EVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILPWNLRYNIALG 452
E+ IS ++HRNLVKL G C + ++ +L+YEY+ N SLD LF L W+ RY+I LG
Sbjct: 182 EIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKALSLNWSTRYDICLG 361
Query: 453 LASALLYLQEEWEKCVIHRDIKSSNIMLDSD 483
+A L YL EE ++HRD+K+SNI+LD +
Sbjct: 362 VARGLTYLHEESRLRIVHRDVKASNILLDHE 454
>BI418434
Length = 478
Score = 140 bits (354), Expect = 5e-34
Identities = 70/157 (44%), Positives = 102/157 (64%)
Frame = +2
Query: 344 ATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHR 403
AT F E K+G GGFG V++G DS S+ A+KR+ G K++ AEV I ++H
Sbjct: 11 ATRGFSE--KVGHGGFGTVFQGELSDS-SLVAVKRLERPGG-GEKEFRAEVSTIGNIQHV 178
Query: 404 NLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILPWNLRYNIALGLASALLYLQEE 463
NLV+L G+C + + +L+YEYM NG+L +L + G L W++R+ +A+G A + YL EE
Sbjct: 179 NLVRLRGFCSENSHRLLVYEYMHNGALSAYLRKDGPCLSWDVRFQVAIGTAKGIAYLHEE 358
Query: 464 WEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHE 500
C++H DIK NI+LDSDF K+ DFGLA+L+ +
Sbjct: 359 LRCCILHCDIKPENILLDSDFTAKVSDFGLAKLIGRD 469
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 139 bits (351), Expect = 1e-33
Identities = 77/204 (37%), Positives = 118/204 (57%), Gaps = 10/204 (4%)
Frame = +3
Query: 302 LWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGG 361
+W+ N E E S + + EF A K S+ +L +AT +F+ +G+GGFG
Sbjct: 324 VWEINT--ESESGSVVNNVKGGSGEFLEVAKLKVFSFGDLKSATKSFKADALIGEGGFGK 497
Query: 362 VYKGYFKD---------SNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWC 412
VYKG+ + S + A+K+++ +S QG ++ +E+ + ++ H NLVKL G+C
Sbjct: 498 VYKGWLDEKKLSPTKPGSGIMVAVKKLNPESMQGFHEWQSEINFLGRISHPNLVKLLGYC 677
Query: 413 HKKNELILIYEYMPNGSLDFHLFRGGS-ILPWNLRYNIALGLASALLYLQEEWEKCVIHR 471
E +L+YE+MP GSL+ HLFR + L WN R IA+G A L +L K VI+R
Sbjct: 678 RDDEEFLLVYEFMPRGSLENHLFRRNTESLSWNTRLKIAIGAARGLAFL-HSLXKIVIYR 854
Query: 472 DIKSSNIMLDSDFNTKLGDFGLAR 495
D K+SNI+LD ++N K+ FGL +
Sbjct: 855 DFKASNILLDGNYNAKISXFGLTK 926
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 139 bits (349), Expect = 2e-33
Identities = 76/182 (41%), Positives = 111/182 (60%), Gaps = 12/182 (6%)
Frame = +3
Query: 393 EVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGG-----------SIL 441
EV+I+S +RH N+VKL + L+L+Y+Y+ N SLD L + +I+
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 442 PWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARL-MDHE 500
W R +IA+G+A L Y+ + V+HRD+K SNI+LDS FN K+ DFGLA + + E
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 501 KGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLV 560
+ + + VAGT GY+APEY T + ++ D++SFGV+LLE+ GK+A H E SL
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGKEANHGDEYS---SLA 533
Query: 561 EW 562
EW
Sbjct: 534 EW 539
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 133 bits (334), Expect = 1e-31
Identities = 75/194 (38%), Positives = 112/194 (57%)
Frame = +1
Query: 324 DDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADS 383
+D+F +G K SY ++ AT NF T LGQG FG VYK + V A+K ++ +S
Sbjct: 163 NDQFASPSGIPKYSYKDIQKATQNF--TTILGQGSFGTVYKATMP-TGEVVAVKVLAPNS 333
Query: 384 RQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILPW 443
+QG ++ EV ++ +L HRNLV L G+C K + IL+Y++M NGSL L+ L W
Sbjct: 334 KQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEKELSW 513
Query: 444 NLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGS 503
+ R IA+ ++ + YL E VIHRD+KS+NI+LD + DFGL++ +
Sbjct: 514 DERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSK--EEIFDG 687
Query: 504 ETTVVAGTRGYLAP 517
+ + GT GY+ P
Sbjct: 688 RNSGLKGTYGYMDP 729
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,056,075
Number of Sequences: 28460
Number of extensions: 179832
Number of successful extensions: 1436
Number of sequences better than 10.0: 307
Number of HSP's better than 10.0 without gapping: 1239
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1244
length of query: 655
length of database: 4,897,600
effective HSP length: 96
effective length of query: 559
effective length of database: 2,165,440
effective search space: 1210480960
effective search space used: 1210480960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144564.8


