

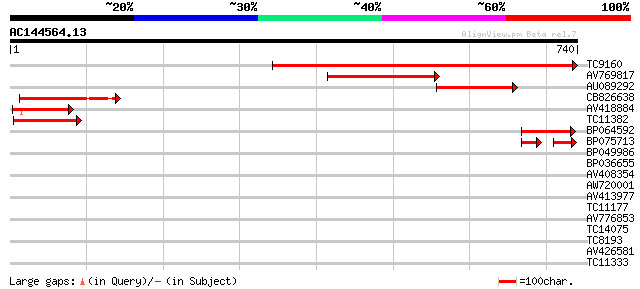
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.13 - phase: 0
(740 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9160 similar to UP|Q8H0V0 (Q8H0V0) Isp4 like protein, partial ... 474 e-134
AV769817 276 1e-74
AU089292 155 2e-38
CB826638 150 7e-37
AV418884 124 5e-29
TC11382 similar to PIR|T01900|T01900 isp4 protein homolog T12H20... 117 9e-27
BP064592 88 4e-18
BP075713 49 4e-13
BP049986 34 0.074
BP036655 30 1.8
AV408354 30 1.8
AW720001 28 4.0
AV413977 28 4.0
TC11177 similar to UP|Q9LTR2 (Q9LTR2) Dbj|BAA17049.1 (AT3g20320/... 28 5.3
AV776853 28 6.9
TC14075 homologue to UP|PRP2_MEDTR (Q40375) Repetitive proline-r... 27 9.0
TC8193 similar to UP|Q9M5J4 (Q9M5J4) Beta-galactosidase (Lactas... 27 9.0
AV426581 27 9.0
TC11333 similar to GB|AAO42855.1|28416649|BT004609 At4g10170 {Ar... 27 9.0
>TC9160 similar to UP|Q8H0V0 (Q8H0V0) Isp4 like protein, partial (53%)
Length = 1554
Score = 474 bits (1220), Expect = e-134
Identities = 215/399 (53%), Positives = 286/399 (70%), Gaps = 1/399 (0%)
Frame = +2
Query: 343 IVNDKFEIDLAKYHEQGRIHLSTFFALSYGFGFATIASTVTHVACFYGREIMERYRASKN 402
I+ +E+++ Y++ G+++LS FALS G GFA +T+THVA F+G +I+ + R++ N
Sbjct: 5 ILTPDYELNVDAYNQYGKLYLSPLFALSIGSGFARFTATLTHVALFHGSDILRQSRSAMN 184
Query: 403 -GKEDIHTKLMKNYKDIPSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFI 461
K D+H +LMK YK +P WWF +LL + +SL++ +Q+PWWG+L A ALAFI
Sbjct: 185 KAKLDVHGRLMKAYKQVPEWWFLILLFGSMALSLVMSFVWKRDVQLPWWGMLFAFALAFI 364
Query: 462 FTLPISIITATTNQTPGLNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKL 521
TLPI +I ATTNQ PG +II +++ G + PG+PIAN+ FK YG IS A+SFLSD KL
Sbjct: 365 VTLPIGVIQATTNQQPGYDIIAQFMIGYVLPGKPIANLLFKIYGRISTVHALSFLSDLKL 544
Query: 522 GHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVF 581
GHYMKIPPR M+ Q +GT++A +N+ VAWW+LD++K+IC D +PWTCP RV
Sbjct: 545 GHYMKIPPRCMYTAQLVGTLVASIVNLAVAWWMLDNIKDICMDDKAHHDNPWTCPKFRVT 724
Query: 582 FDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVL 641
FDASV+WGL+GP+R+FG G Y L W FL GA+ P+ VW+L K FP++ WI LIN+PV+
Sbjct: 725 FDASVIWGLIGPRRLFGPGGLYRNLVWLFLIGAVLPVPVWVLSKIFPEKKWIALINIPVI 904
Query: 642 LGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLY 701
MPPAT N SW++ G IFNFF+FRY ++WWQ+YNYVLSAALDAG AFM VL++
Sbjct: 905 SYGFAGMPPATPSNIASWLVTGMIFNFFVFRYHQRWWQKYNYVLSAALDAGTAFMGVLIF 1084
Query: 702 LALGLENVSLNWWGTAGEHCPLAACPTAKGIVVDGCPVF 740
AL SL WWG+ + CPLA+CPTA GI VDGCPVF
Sbjct: 1085FALPNAGHSLKWWGSEMDPCPLASCPTAPGIKVDGCPVF 1201
>AV769817
Length = 436
Score = 276 bits (705), Expect = 1e-74
Identities = 128/145 (88%), Positives = 137/145 (94%)
Frame = +2
Query: 416 KDIPSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQ 475
+DIPSWWFY LL T +SL++CIFLNDQIQMPWWGLL A ALAF+FTLPISIITATTNQ
Sbjct: 2 EDIPSWWFYALLVATLAISLVLCIFLNDQIQMPWWGLLFAGALAFVFTLPISIITATTNQ 181
Query: 476 TPGLNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLV 535
TPGLNIITEY+FG+IYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLV
Sbjct: 182 TPGLNIITEYVFGLIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLV 361
Query: 536 QFIGTVLAGTINIGVAWWLLDSVKN 560
QFIGTVLAGTINIGVAWWLL+S+ N
Sbjct: 362 QFIGTVLAGTINIGVAWWLLNSIDN 436
>AU089292
Length = 345
Score = 155 bits (393), Expect = 2e-38
Identities = 63/106 (59%), Positives = 84/106 (78%)
Frame = +2
Query: 557 SVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIG 616
++ ++C+ LP PW CP D VF+DASV+WGL+GP+RIFG G Y +N+FFLGGAI
Sbjct: 11 TIPDLCDASKLPTDXPWLCPMDNVFYDASVIWGLLGPRRIFGDQGVYGKVNYFFLGGAIA 190
Query: 617 PILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIV 662
P+LVWL HKAFP + W+ LI++P++LGAT MMPPATA+NY SW++V
Sbjct: 191 PLLVWLAHKAFPXKEWLRLIHMPIMLGATSMMPPATAVNYTSWLLV 328
>CB826638
Length = 575
Score = 150 bits (379), Expect = 7e-37
Identities = 75/131 (57%), Positives = 94/131 (71%)
Frame = +1
Query: 14 KEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLII 73
K++ + PIE+VRLTV TDDPT P TFR W LGL SC LL+F+NQF YRT P+ I
Sbjct: 193 KDQYTVNDCPIEQVRLTVPITDDPTQPALTFRTWVLGLASCVLLAFVNQFLGYRTNPMNI 372
Query: 74 TQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAF 133
T ++ Q+ TLPLG LMA+ LP++ R+P F S FS NPGPF+MKEHVLITIFA++
Sbjct: 373 TSVSAQIVTLPLGKLMAATLPTRPIRVP-FTSWSFSLNPGPFSMKEHVLITIFASS---- 537
Query: 134 GSGSSYAVGIV 144
GSG YA+ I+
Sbjct: 538 GSGGVYAISII 570
>AV418884
Length = 418
Score = 124 bits (311), Expect = 5e-29
Identities = 63/84 (75%), Positives = 70/84 (83%), Gaps = 4/84 (4%)
Frame = +2
Query: 4 INVDTEKGTMK----EEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSF 59
+N DTEK T + +E+E SP+EEVRLTV TDDPT PVWTFRMWFLGLLSC+LLSF
Sbjct: 53 MNSDTEKQTTAATDVDIDEDELSPVEEVRLTVTNTDDPTQPVWTFRMWFLGLLSCSLLSF 232
Query: 60 LNQFFAYRTEPLIITQITVQVATL 83
LNQFFAYRTEPLIIT ITVQV+TL
Sbjct: 233 LNQFFAYRTEPLIITLITVQVSTL 304
>TC11382 similar to PIR|T01900|T01900 isp4 protein homolog T12H20.7 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(10%)
Length = 606
Score = 117 bits (292), Expect = 9e-27
Identities = 54/88 (61%), Positives = 69/88 (78%)
Frame = +3
Query: 6 VDTEKGTMKEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFA 65
V+ E +EEE EEESP+++V LTV KTDDP+L + TFRMWFLG++SC LLSF+NQFF
Sbjct: 342 VNREHEEEEEEEIEEESPVKQVELTVPKTDDPSLHILTFRMWFLGVVSCVLLSFVNQFFW 521
Query: 66 YRTEPLIITQITVQVATLPLGHLMASVL 93
YRTEPL +T I+ Q+A +P+GH MA L
Sbjct: 522 YRTEPLTVTSISAQIAVVPIGHFMARTL 605
>BP064592
Length = 531
Score = 88.2 bits (217), Expect = 4e-18
Identities = 39/72 (54%), Positives = 46/72 (63%), Gaps = 1/72 (1%)
Frame = -2
Query: 668 FFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENV-SLNWWGTAGEHCPLAAC 726
F+++R K WW Y+LSAALDAGVAFM +LLY AL V WWG +HCPLA C
Sbjct: 530 FYVYRNFKXWWXSXTYILSAALDAGVAFMGLLLYFALPSNGVFGPTWWGLDNDHCPLARC 351
Query: 727 PTAKGIVVDGCP 738
PT G+ GCP
Sbjct: 350 PTYPGVYAKGCP 315
>BP075713
Length = 381
Score = 48.5 bits (114), Expect(2) = 4e-13
Identities = 20/26 (76%), Positives = 22/26 (83%)
Frame = -3
Query: 669 FIFRYRKKWWQRYNYVLSAALDAGVA 694
F+FRY WWQ+YNYVLSAALDAG A
Sbjct: 379 FVFRYPHPWWQKYNYVLSAALDAGPA 302
Score = 43.1 bits (100), Expect(2) = 4e-13
Identities = 17/30 (56%), Positives = 20/30 (66%)
Frame = -2
Query: 710 SLNWWGTAGEHCPLAACPTAKGIVVDGCPV 739
SL WWG+ + CPLA+CP G VDGC V
Sbjct: 254 SLKWWGSEMDPCPLASCPLPPGSKVDGCHV 165
>BP049986
Length = 608
Score = 34.3 bits (77), Expect = 0.074
Identities = 23/68 (33%), Positives = 31/68 (44%)
Frame = -2
Query: 11 GTMKEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEP 70
G M+EEEE EE EEV L + K + VW ++ L LL L T P
Sbjct: 490 GRMEEEEEVEEEEEEEVSLAMMKKTRAAI-VWILKLGHLLLLEGLCFGLLEDRIRLFTHP 314
Query: 71 LIITQITV 78
L++ +V
Sbjct: 313 LLLPTSSV 290
>BP036655
Length = 556
Score = 29.6 bits (65), Expect = 1.8
Identities = 17/34 (50%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Frame = -3
Query: 7 DTEKGTMKEEEEEEESPIEEV--RLTVKKTDDPT 38
+TE T KEEEEEEE +E V + VKK + P+
Sbjct: 278 ETEIETEKEEEEEEECGVENVEDEMGVKKFNRPS 177
>AV408354
Length = 428
Score = 29.6 bits (65), Expect = 1.8
Identities = 15/60 (25%), Positives = 28/60 (46%), Gaps = 2/60 (3%)
Frame = +2
Query: 12 TMKEEEEEEESPIEEVRLTVKKTDDPT--LPVWTFRMWFLGLLSCALLSFLNQFFAYRTE 69
T + + E+ E T K+ +P + W+F W+LG +C L + F+ + T+
Sbjct: 128 TNSKHDNMSEAKKVETTQT*KERKNPIY*IGFWSFTQWWLGTWACCRLPVTSSFWRHNTQ 307
>AW720001
Length = 542
Score = 28.5 bits (62), Expect = 4.0
Identities = 27/87 (31%), Positives = 36/87 (41%), Gaps = 12/87 (13%)
Frame = -2
Query: 87 HLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAFGS----------G 136
H S LP+ IPG RFS + NMK + + AFGS
Sbjct: 292 HFSPSTLPTSVNEIPGLVFFRFSLSICEKNMKPDIALL------GAFGSFFFFCFFFFPS 131
Query: 137 SSYAVGIVNIIKAFYGRNI--SFLAAW 161
SS ++G + A GR+ SFLA +
Sbjct: 130 SSASLGFFLEVDALEGRSFFGSFLAGF 50
>AV413977
Length = 397
Score = 28.5 bits (62), Expect = 4.0
Identities = 19/46 (41%), Positives = 26/46 (56%)
Frame = +3
Query: 268 LGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAYW 313
L A +D++AVA L S L+ PF IV V + L+V AV+ W
Sbjct: 57 LPAAIVDYAAVALVLLSLLLPPFLMIVVVLL---LMVAAVVSR*QW 185
>TC11177 similar to UP|Q9LTR2 (Q9LTR2) Dbj|BAA17049.1 (AT3g20320/MQC12_7),
partial (20%)
Length = 601
Score = 28.1 bits (61), Expect = 5.3
Identities = 18/52 (34%), Positives = 22/52 (41%)
Frame = -2
Query: 557 SVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNW 608
S K IC K + W P +F SV+W L K I + Y LNW
Sbjct: 387 SQKTICTSVKTKKNTLWKQPVRNCYFITSVLWLL---KNIGIA*SNYILLNW 241
>AV776853
Length = 625
Score = 27.7 bits (60), Expect = 6.9
Identities = 10/43 (23%), Positives = 22/43 (50%)
Frame = -2
Query: 607 NWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMP 649
NWF + +I P++V + P + +N+P+++ + P
Sbjct: 438 NWFMISFSICPVIVLFTPQPLPIADKLTHVNIPMIISTNIIFP 310
>TC14075 homologue to UP|PRP2_MEDTR (Q40375) Repetitive proline-rich cell
wall protein 2 precursor, partial (98%)
Length = 1524
Score = 27.3 bits (59), Expect = 9.0
Identities = 30/101 (29%), Positives = 40/101 (38%), Gaps = 12/101 (11%)
Frame = -1
Query: 539 GTVLAGTINIG-VAWWLLDSVKNICNKDLLPKGSP------WTCPSDRVFFDASVVW--- 588
G V G +N G V WWLL + N+ LL G W + + V W
Sbjct: 630 GLVNWGFLNWGLVHWWLLH--RRFVNRWLLNWGLVHGWLLNWRLVHRWLLYWGLVHWWFL 457
Query: 589 --GLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAF 627
GLV ++ L + LNW G + WLLH+ F
Sbjct: 456 NWGLVNWGLLYRGLVNWGFLNW-------GLVHWWLLHRRF 355
>TC8193 similar to UP|Q9M5J4 (Q9M5J4) Beta-galactosidase (Lactase) ,
partial (80%)
Length = 2257
Score = 27.3 bits (59), Expect = 9.0
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = -1
Query: 272 TLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVI 308
T DW+ + S SP++SP V +FVG L I
Sbjct: 379 TRDWTTIISVPASPILSPHLGFV-IFVGGELFTVVCI 272
>AV426581
Length = 429
Score = 27.3 bits (59), Expect = 9.0
Identities = 13/30 (43%), Positives = 19/30 (63%)
Frame = -2
Query: 7 DTEKGTMKEEEEEEESPIEEVRLTVKKTDD 36
D+ T++EEEEEEE PI+ + K+ D
Sbjct: 179 DSNLKTVEEEEEEEEDPIKLYKKMQKEVCD 90
>TC11333 similar to GB|AAO42855.1|28416649|BT004609 At4g10170 {Arabidopsis
thaliana;}, partial (14%)
Length = 487
Score = 27.3 bits (59), Expect = 9.0
Identities = 11/31 (35%), Positives = 20/31 (64%)
Frame = +1
Query: 674 RKKWWQRYNYVLSAALDAGVAFMAVLLYLAL 704
R+KWW++ VL A+DA V + +++L +
Sbjct: 64 RRKWWRQVRIVL--AIDAAVCIILFVIWLVI 150
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,022,742
Number of Sequences: 28460
Number of extensions: 241964
Number of successful extensions: 2137
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1954
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2069
length of query: 740
length of database: 4,897,600
effective HSP length: 97
effective length of query: 643
effective length of database: 2,136,980
effective search space: 1374078140
effective search space used: 1374078140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144564.13


