

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.2 + phase: 0
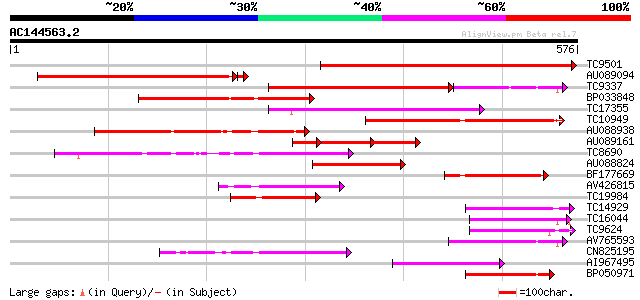
(576 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesteras... 413 e-116
AU089094 276 4e-75
TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B pre... 176 7e-66
BP033848 230 5e-61
TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%) 191 3e-49
TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precu... 182 2e-46
AU088938 176 9e-45
AU089161 71 1e-33
TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precu... 124 3e-29
AU088824 104 4e-23
BF177669 100 6e-22
AV426815 99 2e-21
TC19984 similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase , pa... 96 2e-20
TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase ... 93 1e-19
TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 91 7e-19
TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precur... 90 9e-19
AV765593 89 2e-18
CN825195 89 3e-18
AI967495 89 3e-18
BP050971 86 2e-17
>TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase ,
partial (29%)
Length = 934
Score = 413 bits (1062), Expect = e-116
Identities = 191/260 (73%), Positives = 223/260 (85%)
Frame = +2
Query: 316 PARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSA 375
PA+TI+TGRK+FA GVKTMQTATFAN A GFI KA+TFENTAGP+GHQAVA RNQGD SA
Sbjct: 5 PAKTIITGRKNFADGVKTMQTATFANQAPGFIAKAITFENTAGPNGHQAVALRNQGDQSA 184
Query: 376 LVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTI 435
VGCHI+GYQD+LYVQ+NRQ+YRNC++SGT+DFIFG+S TLIQHS I+VRKP QFNT+
Sbjct: 185 FVGCHILGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSPTLIQHSIIVVRKPNDNQFNTV 364
Query: 436 TADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIH 495
TADG+ T N+ TGIVIQDC I+PEAALFP RF I+SYLGRPWK ++TVVMEST+GDFIH
Sbjct: 365 TADGTPTKNMPTGIVIQDCEIVPEAALFPVRFKIKSYLGRPWKEYSRTVVMESTLGDFIH 544
Query: 496 PDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG 555
P+GW W G +T YYAE+ANTGPGAN A R+KWKGY GVISRAEA +FTAG +L+AG
Sbjct: 545 PEGWFPWAGSFALDTLYYAEHANTGPGANTAGRIKWKGYRGVISRAEATQFTAGEFLKAG 724
Query: 556 PKSAAEWLNGLHVPHYLGFK 575
P+S +W+ LHVPHYLGFK
Sbjct: 725 PRSGLDWIKALHVPHYLGFK 784
>AU089094
Length = 639
Score = 276 bits (706), Expect(2) = 4e-75
Identities = 138/204 (67%), Positives = 166/204 (80%), Gaps = 1/204 (0%)
Frame = +1
Query: 29 NGEDPEVQTQQRNLRIMCQNSQDQKLCHETLSSVHGADAADPKAYIAASVKAATDNVIKA 88
N EVQTQQR++R +CQN+ DQKLCH+TLSSV G DAADPKAYIA +VKAA+D+VIKA
Sbjct: 1 NSAPAEVQTQQRSVRTICQNTDDQKLCHDTLSSVKGVDAADPKAYIATAVKAASDSVIKA 180
Query: 89 FNMSERLTTEYG-KENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRN 147
FNMS+RLTTEYG K++G KMAL+DCKDL+Q A+ SLDLST V DNNIQ VH+QI D RN
Sbjct: 181 FNMSDRLTTEYGNKDHGIKMALDDCKDLLQSAMQSLDLSTTLVRDNNIQGVHNQIPDFRN 360
Query: 148 WLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFN 207
WL AVISY+QACMEGFD+ DGEK IKEQ +S+D V ++ A+ LDIVT ++ IL QFN
Sbjct: 361 WLGAVISYQQACMEGFDNDXDGEKXIKEQLXTESMDHVXRIXAITLDIVTNMTKILXQFN 540
Query: 208 LKFDVKPASRRLLNSEVTVDDQGY 231
LK D+KPASRRLL + VD++GY
Sbjct: 541 LKLDLKPASRRLLXXD--VDNEGY 606
Score = 22.7 bits (47), Expect(2) = 4e-75
Identities = 8/11 (72%), Positives = 10/11 (90%)
Frame = +3
Query: 232 PSWISSSDRKL 242
PSW S+S+RKL
Sbjct: 606 PSWFSASERKL 638
>TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (54%)
Length = 1145
Score = 176 bits (445), Expect(2) = 7e-66
Identities = 85/188 (45%), Positives = 120/188 (63%)
Frame = +2
Query: 264 DGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTG 323
DGSG + T+ A+ + P + R+VIYVK GVY+EY + K NI+M GDG T+++G
Sbjct: 29 DGSGNYTTVMEAVLAAPDYSMKRFVIYVKKGVYEEYAEIKKKKWNIMMVGDGMDVTVISG 208
Query: 324 RKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVG 383
+SF G T ++ATFA + GFI + +TF+NTAGP+ HQAVA R+ D++ C I G
Sbjct: 209 NRSFGDGWTTFKSATFAVSGRGFIARGITFQNTAGPEKHQAVALRSDSDLAVFFRCSITG 388
Query: 384 YQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTM 443
YQDSLY + RQ+Y+ C ++GTVDFIFG + + Q+ I RK + Q NTITA G
Sbjct: 389 YQDSLYPHTMRQFYKECRITGTVDFIFGDATAVFQNCNIQARKGLQDQKNTITAQGRKDP 568
Query: 444 NLNTGIVI 451
++G +
Sbjct: 569 GQSSGFFV 592
Score = 92.4 bits (228), Expect(2) = 7e-66
Identities = 47/118 (39%), Positives = 68/118 (56%), Gaps = 3/118 (2%)
Frame = +3
Query: 452 QDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTC 511
Q CNI ++ L P +I++YLGRPWK ++TV M+S + D + GW W + +T
Sbjct: 594 QYCNITADSELLPSVGSIQTYLGRPWKEYSRTVFMQSFMSDVVSSQGWLEWNTSFD-DTL 770
Query: 512 YYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG---PKSAAEWLNGL 566
+Y EY N G GA V RVKW GYH V++ +EA KFT +++ P + + GL
Sbjct: 771 FYGEYLNYGAGAGVGNRVKWGGYH-VLNDSEAGKFTVNQFIEGNLWLPSTGVAYTGGL 941
>BP033848
Length = 525
Score = 230 bits (586), Expect = 5e-61
Identities = 115/178 (64%), Positives = 140/178 (78%)
Frame = +2
Query: 132 DNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAV 191
DNNIQAVH+Q D RNWLSAV+SY+Q+CMEGF+D GE+ +K Q SLD + K+T +
Sbjct: 2 DNNIQAVHEQTPDFRNWLSAVMSYQQSCMEGFEDGKSGEEAVKGQLQEGSLDQMGKLTGI 181
Query: 192 ALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNW 251
ALDIV+ LS ILQ F+LK D+ PASRRLL +E VD +G PSW S +DRKLL KM++
Sbjct: 182 ALDIVSDLSRILQTFDLKLDLNPASRRLLEAE-EVDHEGLPSWFSGADRKLLGKMKK--- 349
Query: 252 RANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNI 309
+ PNAVVAKDGSG+FKTI +A+ASYPKG KGRY+IYV+AGVYDEYITVPK AVN+
Sbjct: 350 GGKVTPNAVVAKDGSGKFKTIASAIASYPKGFKGRYIIYVRAGVYDEYITVPKQAVNL 523
>TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%)
Length = 848
Score = 191 bits (485), Expect = 3e-49
Identities = 103/222 (46%), Positives = 134/222 (59%), Gaps = 3/222 (1%)
Frame = +1
Query: 264 DGSGQFKTIQAALASYPKGNK---GRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTI 320
+G+ + +I A+A+ P K G ++IYV+ G Y+EY+ VPK+ NIL+ GDG +TI
Sbjct: 172 NGTDNYTSIGDAIAAAPNHTKPQDGYFLIYVREGYYEEYVIVPKEKKNILLVGDGINKTI 351
Query: 321 VTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCH 380
+TG S G T ++TFA + F +TF NTAGP HQAVA RN D+S C
Sbjct: 352 ITGNHSVIDGWTTFNSSTFAVSGERFTAVDVTFRNTAGPAKHQAVAVRNNADLSTFYRCS 531
Query: 381 IVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGS 440
GYQD+LYV S RQ+YR C + GTVDFIFG++A + Q+ I RKP Q N +TA G
Sbjct: 532 FEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAAVVFQNCNIYARKPLPFQKNAVTAQGR 711
Query: 441 DTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAK 482
N NTGI IQ+C I L E + SYLGRPW+ L K
Sbjct: 712 TDPNQNTGISIQNCTIDAAPDLAAELNSTLSYLGRPWEGLFK 837
>TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (34%)
Length = 773
Score = 182 bits (461), Expect = 2e-46
Identities = 93/203 (45%), Positives = 126/203 (61%), Gaps = 1/203 (0%)
Frame = +3
Query: 362 HQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHST 421
HQAVA R+ D S C G+QD+LY SNRQ+YR C ++GT+DFIFG++A++ Q+
Sbjct: 9 HQAVAMRSGSDRSVFYRCSFDGFQDTLYAHSNRQFYRECDITGTIDFIFGNAASIFQNCK 188
Query: 422 IIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLA 481
I+ R+P QF TITA G N NTGIVIQ I P +T +YLGRPWK +
Sbjct: 189 IMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITP----LESTYTAPTYLGRPWKDFS 356
Query: 482 KTVVMESTIGDFIHPDGWTIWQGEQN-HNTCYYAEYANTGPGANVARRVKWKGYHGVISR 540
T++M+S IG F+ P GW W + T +YAE+ N+GPG++VA+RVKW GY ++
Sbjct: 357 TTLIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTA 536
Query: 541 AEANKFTAGIWLQAGPKSAAEWL 563
+ KFT ++Q GP EWL
Sbjct: 537 DDLGKFTLASFIQ-GP----EWL 590
>AU088938
Length = 641
Score = 176 bits (446), Expect = 9e-45
Identities = 98/219 (44%), Positives = 142/219 (64%), Gaps = 1/219 (0%)
Frame = +1
Query: 87 KAFNMSERLTTEYGKEN-GAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADM 145
+AFNMS+RLT E+ N G KMAL DCKDL+Q A+ L S V + V+ + A++
Sbjct: 7 EAFNMSDRLTVEHENSNPGVKMALEDCKDLLQSAIQELQASGVLVQXXXLLDVNQRSAEL 186
Query: 146 RNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQ 205
+NWLSAV++Y+Q+C++GFD GEK ++++ SLD V K+T +ALDIV+G+S +L
Sbjct: 187 KNWLSAVVAYQQSCLDGFD--TKGEKXVQDKLQQGSLDDVGKLTGLALDIVSGISKVLGA 360
Query: 206 FNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDG 265
F+ +KPA RRLL +D +GYP+W+S +DRKL+ ++N++PNA+VAKDG
Sbjct: 361 FDFNLHLKPA-RRLLE----IDAEGYPTWLSFADRKLVES------KSNVLPNAIVAKDG 507
Query: 266 SGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPK 304
G FK I A+ SY + +AGVYD+Y T+ K
Sbjct: 508 XGXFKNITEAINSYLXXTXVNPHL-CQAGVYDKYSTLTK 621
>AU089161
Length = 400
Score = 70.9 bits (172), Expect(3) = 1e-33
Identities = 33/59 (55%), Positives = 41/59 (68%)
Frame = +1
Query: 313 GDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQG 371
G GP +T++TG K+F G KTM++ATF A F+ K+M FENTAG HQAVA R QG
Sbjct: 85 GXGPKKTVITGNKNFVDGYKTMRSATFTTVAPDFMAKSMGFENTAGASKHQAVALRVQG 261
Score = 64.7 bits (156), Expect(3) = 1e-33
Identities = 30/51 (58%), Positives = 36/51 (69%)
Frame = +2
Query: 367 FRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLI 417
F + D SA C I GYQD+LY ++RQ+YRNC +SGTVDFIFG S LI
Sbjct: 248 FACKXDRSAFFDCAISGYQDTLYAHAHRQFYRNCEISGTVDFIFGYSTLLI 400
Score = 45.1 bits (105), Expect(3) = 1e-33
Identities = 21/29 (72%), Positives = 22/29 (75%)
Frame = +3
Query: 288 VIYVKAGVYDEYITVPKDAVNILMYGDGP 316
VIYVKAGVYDEYI + K NILMYG P
Sbjct: 9 VIYVKAGVYDEYIQIDKKKPNILMYGXWP 95
>TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor
(Pectinesterase) , partial (59%)
Length = 1082
Score = 124 bits (312), Expect = 3e-29
Identities = 91/307 (29%), Positives = 149/307 (47%), Gaps = 3/307 (0%)
Frame = +3
Query: 46 CQNSQDQKLCHETLSSVHGADAA---DPKAYIAASVKAATDNVIKAFNMSERLTTEYGKE 102
C+++ D C +S V + + DPK I S+ + + I+ + +
Sbjct: 243 CEHALDASSCLAHVSEVSQSPISATKDPKLNILISLMTKSTSHIQEAMVKTKAIKNRINN 422
Query: 103 NGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEG 162
+ AL+DC+ LM ++D + S + +N D D WLS V++ C++G
Sbjct: 423 PREEAALSDCEQLMDLSIDRVWDSVMALTKDNT----DSHQDAHAWLSGVLTNHATCLDG 590
Query: 163 FDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNS 222
+ G + + ++ L S K T++AL +V+ L+ K + ++++
Sbjct: 591 LE----GPSRALMEAEIEDLISRSK-TSLAL-LVSVLAP-----------KGGNEQIIDE 719
Query: 223 EVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKG 282
+ D +PSW++ DR+LL ++ N VVAKDGSG+FKT+ A+AS P
Sbjct: 720 PL---DGDFPSWVTRKDRRLLESSV-----GDVNANVVVAKDGSGRFKTVAEAVASAPDS 875
Query: 283 NKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANT 342
K RYVIYVK G Y E I + K N+++ GDG TI+TG + G T ++AT A
Sbjct: 876 GKTRYVIYVKKGTYKENIEIGKKKTNVMLTGDGMDATIITGNLNVIDGSTTFKSATVAAV 1055
Query: 343 AMGFIGK 349
GFI +
Sbjct: 1056GDGFIAQ 1076
>AU088824
Length = 421
Score = 104 bits (259), Expect = 4e-23
Identities = 54/95 (56%), Positives = 64/95 (66%)
Frame = +1
Query: 308 NILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAF 367
NILMYGDGP +TI+TGRK+FA GVKTMQTATFAN A GFI KA+TFENTAGP +
Sbjct: 10 NILMYGDGPTKTIITGRKNFADGVKTMQTATFANQAPGFIAKAITFENTAGPTVTKLWH* 189
Query: 368 RNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLV 402
+G SA +G L VQ+ Q+YR C +
Sbjct: 190 XTKGXQSAFRWAAHLGLPKXLLVQTTXQFYRXCFL 294
>BF177669
Length = 336
Score = 100 bits (249), Expect = 6e-22
Identities = 50/106 (47%), Positives = 67/106 (63%)
Frame = +3
Query: 442 TMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTI 501
T+ GI IQ+C+I P F +++YLGRPWK + TVVM+ST+G FI P+GW
Sbjct: 24 TLT*TQGISIQNCSISP----FGNLSYVQTYLGRPWKNYSTTVVMQSTLGSFISPNGWLP 191
Query: 502 WQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFT 547
W G +T +YAEY N G GA+ RVKWKG + S+ +A+KFT
Sbjct: 192 WVGNSAPDTIFYAEYQNVGQGASTKNRVKWKGLKTITSK-QASKFT 326
>AV426815
Length = 391
Score = 98.6 bits (244), Expect = 2e-21
Identities = 54/128 (42%), Positives = 76/128 (59%)
Frame = +3
Query: 213 KPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTI 272
K SR+L+ + + +P WIS++DR+LL + + VVA DGSG FKT+
Sbjct: 24 KTGSRKLMEA-----GEVWPEWISAADRRLLQA-------GTVKADVVVAADGSGNFKTV 167
Query: 273 QAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVK 332
AA+A+ P+ +K RYVI +KAGVY E + VPK NI+ GDG TI+TG ++ G
Sbjct: 168 SAAVAAAPEKSKTRYVIKIKAGVYRENVEVPKKKTNIMFLGDGRTTTIITGSRNVVDGST 347
Query: 333 TMQTATFA 340
T +AT A
Sbjct: 348 TFHSATVA 371
>TC19984 similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase , partial (11%)
Length = 300
Score = 95.5 bits (236), Expect = 2e-20
Identities = 47/91 (51%), Positives = 56/91 (60%)
Frame = +3
Query: 225 TVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNK 284
++D G+PSW+ DR+LL + PN VAKDGSG FKTI AL P K
Sbjct: 33 SLDSDGFPSWVHVDDRRLLKAADNRP-----KPNVTVAKDGSGDFKTISEALNKIPATYK 197
Query: 285 GRYVIYVKAGVYDEYITVPKDAVNILMYGDG 315
GRYV+YVK GVYDE +TV K N+ MYGDG
Sbjct: 198 GRYVVYVKEGVYDEQVTVTKKMQNVTMYGDG 290
>TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase (Fragment)
, partial (24%)
Length = 693
Score = 92.8 bits (229), Expect = 1e-19
Identities = 45/110 (40%), Positives = 61/110 (54%)
Frame = +2
Query: 464 PERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGA 523
P +++ +YLGRPWK ++TV M+S+I D I P GW W G NT Y EY GPGA
Sbjct: 5 PVKYSFPTYLGRPWKEYSRTVFMQSSISDVIDPAGWHEWSGNFALNTLVYREYQYPGPGA 184
Query: 524 NVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
++RV WKG + S +EA FT G ++ + + WL P LG
Sbjct: 185 GTSKRVAWKGVKVITSASEAQAFTPGSFI-----AGSNWLGSTGFPFSLG 319
>TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (43%)
Length = 564
Score = 90.5 bits (223), Expect = 7e-19
Identities = 45/109 (41%), Positives = 61/109 (55%), Gaps = 6/109 (5%)
Frame = +3
Query: 468 TIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVAR 527
+ +YLGRPWK ++TV M S IGD +H GW W +T YY EY N GPGA V +
Sbjct: 21 SFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQ 200
Query: 528 RVKWKGYHGVISRAEANKFTAGIWLQAG---PKSAAEWLNGLH---VPH 570
RV W GY + S EA++FT ++ P + +L+GL +PH
Sbjct: 201 RVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGLST*IIPH 347
>TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precursor
(Pectinesterase) (Pectin methylesterase) (Fragment) ,
partial (33%)
Length = 586
Score = 90.1 bits (222), Expect = 9e-19
Identities = 44/115 (38%), Positives = 66/115 (57%), Gaps = 8/115 (6%)
Frame = +3
Query: 468 TIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNH--NTCYYAEYANTGPGANV 525
+I++YLGRPWK ++T+V++S+I I P GW W + T YY EY N+G GA
Sbjct: 9 SIKTYLGRPWKKYSRTIVLQSSIDSHIDPTGWAEWDAQSKDFLQTLYYGEYLNSGAGAGT 188
Query: 526 ARRVKWKGYHGVISRAEANKFT------AGIWLQAGPKSAAEWLNGLHVPHYLGF 574
+RV W G+H + + AEA+KFT +WL+ ++ GL + Y+GF
Sbjct: 189 GKRVNWPGFHVIKTAAEASKFTVAQLIQGNVWLKG---KGVNFIEGL*I--YIGF 338
>AV765593
Length = 501
Score = 89.0 bits (219), Expect = 2e-18
Identities = 47/124 (37%), Positives = 68/124 (53%), Gaps = 3/124 (2%)
Frame = -1
Query: 446 NTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGE 505
+TG I +C I L +YLGRPWK ++TV M++++ + I GW W GE
Sbjct: 498 DTGTSIHNCTIRAADDLASSNGATATYLGRPWKEYSRTVYMQTSMDNVIDSAGWRAWDGE 319
Query: 506 QNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAG---PKSAAEW 562
+T YYAE+ N+GPG+N RV W+GYH VI+ +A FT +L P++ +
Sbjct: 318 FALSTLYYAEFNNSGPGSNTNGRVTWQGYH-VINATDAANFTVSNFLLGDDWLPQTGVSY 142
Query: 563 LNGL 566
N L
Sbjct: 141 TNTL 130
>CN825195
Length = 551
Score = 88.6 bits (218), Expect = 3e-18
Identities = 59/197 (29%), Positives = 104/197 (51%), Gaps = 2/197 (1%)
Frame = +2
Query: 153 ISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDV 212
++ + C +GF D + +K+Q V +L + ++ + L I G D ++
Sbjct: 17 LTNQDTCADGFADTSGD---VKDQMAV-NLKDLSELVSNCLAIFAGAGDDFSGVPIQ--- 175
Query: 213 KPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTI 272
+RRLL + D +P W++ DR+LL+ + + + VV+KDG+G KTI
Sbjct: 176 ---NRRLL----AMRDDNFPRWLNRRDRRLLSLPV-----SAMQADVVVSKDGNGTAKTI 319
Query: 273 QAALASYPKGNKGRYVIYVKAGVYDE-YITVPKDAVNILMYGDGPARTIVTGRKSFAAG- 330
A+ P+ R++IYV+AG Y+E + + + N+++ GDG +T++TG K+ G
Sbjct: 320 AEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMYGL 499
Query: 331 VKTMQTATFANTAMGFI 347
+ T TA+FA + GFI
Sbjct: 500 LTTFHTASFAASGAGFI 550
>AI967495
Length = 390
Score = 88.6 bits (218), Expect = 3e-18
Identities = 44/113 (38%), Positives = 64/113 (55%)
Frame = +1
Query: 390 VQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGI 449
V + RQ+Y+ C + GTVD IFG++A + Q+ I +KP GQ N ITA G D NTGI
Sbjct: 16 VHAQRQFYKQCQIYGTVDLIFGNAAVVFQNCKIFAKKPLDGQANMITAQGRDDPFQNTGI 195
Query: 450 VIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIW 502
I + I L P + LGRPW+ ++ VV+++ + + P GW+ W
Sbjct: 196 TIHNSEIRAAPDLKPVVNKHNTSLGRPWQQYSRVVVLKTFMDTLVSPLGWSSW 354
>BP050971
Length = 477
Score = 85.5 bits (210), Expect = 2e-17
Identities = 38/91 (41%), Positives = 59/91 (64%), Gaps = 1/91 (1%)
Frame = -1
Query: 464 PERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNH-NTCYYAEYANTGPG 522
P +I ++LGRPWK ++TV MES + + IHP+GW W+ + + ++ EY N GPG
Sbjct: 462 PSNGSIPTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETYTTYDDPLFFGEYPNKGPG 283
Query: 523 ANVARRVKWKGYHGVISRAEANKFTAGIWLQ 553
+ VA RVKW+GYH V++ + A FT +++
Sbjct: 282 SGVANRVKWRGYH-VLNNSPAMDFTVNPFIK 193
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,977,906
Number of Sequences: 28460
Number of extensions: 111041
Number of successful extensions: 522
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 497
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 500
length of query: 576
length of database: 4,897,600
effective HSP length: 95
effective length of query: 481
effective length of database: 2,193,900
effective search space: 1055265900
effective search space used: 1055265900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144563.2


