

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.11 - phase: 0
(322 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
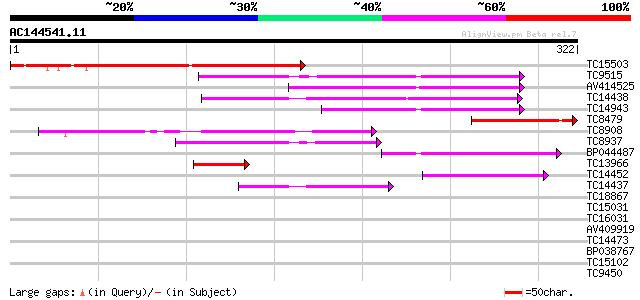
Score E
Sequences producing significant alignments: (bits) Value
TC15503 similar to UP|O04018 (O04018) F7G19.1 protein, partial (... 219 4e-58
TC9515 similar to UP|O49081 (O49081) Clp-like energy-dependent p... 101 1e-22
AV414525 89 1e-18
TC14438 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteo... 80 4e-16
TC14943 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protea... 79 1e-15
TC8479 similar to UP|O04018 (O04018) F7G19.1 protein, partial (9%) 79 1e-15
TC8908 similar to UP|Q8LAN0 (Q8LAN0) ATP-dependent Clp protease ... 69 1e-12
TC8937 similar to UP|Q8W4Z1 (Q8W4Z1) ClpP (ATP-dependent Clp pr... 64 4e-11
BP044487 62 1e-10
TC13966 homologue to UP|O23548 (O23548) CLP proteinase homolog (... 47 6e-06
TC14452 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease... 44 5e-05
TC14437 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteo... 43 8e-05
TC18867 similar to UP|Q9ASV2 (Q9ASV2) At1g06730/F4H5_22, partial... 35 0.022
TC15031 weakly similar to UP|Q69107 (Q69107) LAT protein, partia... 34 0.038
TC16031 similar to UP|O23548 (O23548) CLP proteinase homolog (CL... 33 0.065
AV409919 33 0.085
TC14473 similar to UP|Q8H1Y2 (Q8H1Y2) 20S proteasome alpha 6 sub... 32 0.14
BP038767 31 0.25
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 31 0.25
TC9450 similar to GB|AAM16172.1|20334832|AY094016 At2g39720/T5I7... 30 0.42
>TC15503 similar to UP|O04018 (O04018) F7G19.1 protein, partial (45%)
Length = 590
Score = 219 bits (559), Expect = 4e-58
Identities = 122/181 (67%), Positives = 143/181 (78%), Gaps = 13/181 (7%)
Frame = +1
Query: 1 MATCFRVPISMPTSMPASST-----RFTPTR-----LRTLRTVNAISKSKIP---FNPND 47
MAT RVP ++PTS+P SS+ + P+R LR+ +VNA +++KIP NPND
Sbjct: 52 MATYLRVP-AVPTSIPTSSSSSNSQQSLPSRRRCRNLRSFCSVNA-TRAKIPTPHLNPND 225
Query: 48 PFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRP 107
PFLSKLASVA SSPE L N + D P+LDIFDSP LMATPAQ+ERS SY++ R P+RP
Sbjct: 226 PFLSKLASVAASSPEMLLNRPVNSDTPPYLDIFDSPTLMATPAQVERSVSYNEHR-PRRP 402
Query: 108 PPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGET 167
PPDLPSLLL+GRIVYIGMPLVPAVTEL++AELM+LQWM PKEPIYIYINSTGTTR DGET
Sbjct: 403 PPDLPSLLLHGRIVYIGMPLVPAVTELIVAELMYLQWMDPKEPIYIYINSTGTTRDDGET 582
Query: 168 V 168
V
Sbjct: 583 V 585
>TC9515 similar to UP|O49081 (O49081) Clp-like energy-dependent protease
(ATP-dependent Clp protease proteolytic subunit)
(Endopeptidase Clp) , partial (74%)
Length = 1158
Score = 101 bits (252), Expect = 1e-22
Identities = 64/185 (34%), Positives = 94/185 (50%)
Frame = +1
Query: 108 PPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGET 167
P DL S+L RI++IG P+ V + VI++L+ L + P I +YIN G G T
Sbjct: 304 PLDLSSVLFRNRIIFIGQPVNAQVAQRVISQLVTLATIDPNADILMYINCPG-----GST 468
Query: 168 VAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRV 227
++ AIYD M +K ++ TV G A Q LLL+ G KG RY P+++ MI QP+
Sbjct: 469 YSV----LAIYDCMSWIKPKVGTVCFGVAASQGALLLAGGEKGMRYSMPNSRIMIHQPQS 636
Query: 228 PSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGV 287
G DV E + +R + K+ + T E V +R ++ A EFG+
Sbjct: 637 GCGG--HVEDVRRQVNEAVQSRHKIDKMYSAFTGQPIEKVQQYTERDRFLSVSEALEFGL 810
Query: 288 IDKIL 292
ID +L
Sbjct: 811 IDGVL 825
>AV414525
Length = 427
Score = 89.0 bits (219), Expect = 1e-18
Identities = 52/134 (38%), Positives = 75/134 (55%)
Frame = +3
Query: 159 GTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHA 218
GT ETV E+E ++I D M +K ++ TV G A GQA +LLS G KG R + P++
Sbjct: 3 GTQNEKNETVGSETEAYSIADMMSYVKADVYTVNCGMAYGQAAMLLSLGAKGYRALQPNS 182
Query: 219 KAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMD 278
+ P+V S A D+ I AKE+ N D ++LLAK S+E V+ ++R Y
Sbjct: 183 STKLYLPKVNRSS-GAAIDMWIKAKELEANSDYYIELLAKGIGKSKEEVAKDVQRPRYFQ 359
Query: 279 ALLAKEFGVIDKIL 292
A E+G+ DKI+
Sbjct: 360 PKEAIEYGIADKII 401
>TC14438 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteolytic
subunit (Endopeptidase Clp) , complete
Length = 620
Score = 80.5 bits (197), Expect = 4e-16
Identities = 47/182 (25%), Positives = 91/182 (49%)
Frame = +3
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ + L R++++G + ++ +I +++L K+ +Y++INS G
Sbjct: 93 DIYNRLYRERLLFLGQEVNSEISNQLIGLMVYLSIEDDKKDLYLFINSPG---------G 245
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M ++ ++ TV +G A +L+ G +R PHA+ MI QP S
Sbjct: 246 WVIPGIAIYDTMQFVQPDVQTVCMGLAASMGSFVLAGGKITKRLAFPHARVMIHQP-ASS 422
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
+ ++ A+E++ R+T+ ++ + T VS M+R +M A A+ +G++D
Sbjct: 423 FYEAQTGEFILEAEELLKLRETITRVYVQRTGKPLWLVSEDMERDVFMSAAEAQAYGIVD 602
Query: 290 KI 291
+
Sbjct: 603 LV 608
>TC14943 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (ATP-dependent Clp protease proteolytic subunit
ClpP5) (Endopeptidase Clp) , partial (42%)
Length = 639
Score = 79.0 bits (193), Expect = 1e-15
Identities = 41/115 (35%), Positives = 68/115 (58%)
Frame = +1
Query: 178 YDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASD 237
+D M ++ +++TV +G A LLSAGTKG+RY P+++ MI QP + G +D
Sbjct: 1 FDTMRHIRPDVSTVCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGAQG--GQTD 174
Query: 238 VLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
+ I A E++ ++ L L+ HT S E ++ R ++M A AKE+G+ID ++
Sbjct: 175 IDIQANEMLHHKANLNGYLSYHTGQSLEKINQDTDRDFFMSAKEAKEYGLIDGVI 339
>TC8479 similar to UP|O04018 (O04018) F7G19.1 protein, partial (9%)
Length = 544
Score = 78.6 bits (192), Expect = 1e-15
Identities = 39/60 (65%), Positives = 49/60 (81%)
Frame = +1
Query: 263 SEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIMADVPSRDDRENGTAGAKVENRF 322
+EETV+NVMKR YYMD+ AKEFGVID+ILWRGQEK+MADV + ++ + G AG KV + F
Sbjct: 1 AEETVANVMKRPYYMDSTRAKEFGVIDQILWRGQEKVMADVAAPEEWDKG-AGIKVVDGF 177
>TC8908 similar to UP|Q8LAN0 (Q8LAN0) ATP-dependent Clp protease
proteolytic subunit (ClpP4), partial (35%)
Length = 598
Score = 68.9 bits (167), Expect = 1e-12
Identities = 58/198 (29%), Positives = 86/198 (43%), Gaps = 6/198 (3%)
Frame = +1
Query: 17 ASSTRFTPTRLRTL------RTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSS 70
+SS F+P+R+RTL + N S P + P L + S F P S
Sbjct: 79 SSSLPFSPSRIRTLFSKPPTSSFNPKPSSPSPCHRTTPLKCILTASPNHSKNANFEPESL 258
Query: 71 PDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPA 130
+ F SPQ TPA R A D LLL RIV++G +
Sbjct: 259 KPRITFQ--MSSPQ---TPATAIRGAE-----------TDAMGLLLRERIVFLGSGIDDF 390
Query: 131 VTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINT 190
V + ++++L+ L P + I ++INS G + S AIYDA ++ ++NT
Sbjct: 391 VADAIMSQLLLLDAQDPSKDIKLFINSPGGSL---------SATMAIYDADQLVRADVNT 543
Query: 191 VALGAAVGQACLLLSAGT 208
+ALG A ++L GT
Sbjct: 544 IALGIAASTDSIILGGGT 597
>TC8937 similar to UP|Q8W4Z1 (Q8W4Z1) ClpP (ATP-dependent Clp protease
proteolytic subunit) (Endopeptidase Clp) (Fragment) ,
partial (40%)
Length = 568
Score = 63.5 bits (153), Expect = 4e-11
Identities = 40/117 (34%), Positives = 60/117 (51%)
Frame = +3
Query: 95 SASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIY 154
SAS + PK D ++LL RIV++G + + +I++L+FL K+ I ++
Sbjct: 243 SASTASPWLPKLEELDTTNMLLRQRIVFLGSQVDDMTADFIISQLLFLDAEDSKKDIKLF 422
Query: 155 INSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGR 211
INS G G A G IYDAM K ++ T LG A +L++GTKG+
Sbjct: 423 INSPG-----GSVTA----GMGIYDAMKLCKADVPTGRLGLAASMGAFILASGTKGK 566
>BP044487
Length = 533
Score = 62.4 bits (150), Expect = 1e-10
Identities = 36/102 (35%), Positives = 55/102 (53%)
Frame = -1
Query: 212 RYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVM 271
R P+A MI QP SG A D+ IH K+++ D L L AKHT S E + M
Sbjct: 533 RRSLPNASIMIHQPSGGYSG--QAKDIAIHTKQIVRVWDLLNDLYAKHTGQSIELIQKNM 360
Query: 272 KRSYYMDALLAKEFGVIDKILWRGQEKIMADVPSRDDRENGT 313
R Y+M AKEFG+ID+++ + +++D + ++ G+
Sbjct: 359 DRDYFMTPEEAKEFGLIDEVIDQRPMTLVSDAVGNEGKDKGS 234
>TC13966 homologue to UP|O23548 (O23548) CLP proteinase homolog (CLP
proteinase like protein), partial (20%)
Length = 417
Score = 46.6 bits (109), Expect = 6e-06
Identities = 20/32 (62%), Positives = 26/32 (80%)
Frame = +3
Query: 105 KRPPPDLPSLLLNGRIVYIGMPLVPAVTELVI 136
++PPPDL S L RIVY+GM LVP+VTEL++
Sbjct: 321 EQPPPDLASYLYKNRIVYLGMSLVPSVTELIL 416
>TC14452 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease-like
protein (ATP-dependent Clp protease proteolytic
subunit) (Endopeptidase Clp) (At5g45390) , partial (36%)
Length = 621
Score = 43.5 bits (101), Expect = 5e-05
Identities = 24/72 (33%), Positives = 40/72 (55%)
Frame = +2
Query: 235 ASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILWR 294
A DV I AKEVM N++ + ++++ T + E V + R YM + A E+G+ID ++ +
Sbjct: 17 AIDVEIQAKEVMHNKNNVTRIISGFTGRTFEQVQKDIDRDRYMSPIEAVEYGIIDGVIDK 196
Query: 295 GQEKIMADVPSR 306
+ VP R
Sbjct: 197 ESIIPLEPVPER 232
>TC14437 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteolytic
subunit (Endopeptidase Clp) , partial (58%)
Length = 1013
Score = 42.7 bits (99), Expect = 8e-05
Identities = 24/88 (27%), Positives = 42/88 (47%)
Frame = +2
Query: 131 VTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINT 190
++ +I +++L K+ +Y++INS G G AIYD M ++ ++ T
Sbjct: 5 ISNQLIGLMVYLSIEDDKKDLYLFINSPG---------GWVIPGIAIYDTMQFVQPDVQT 157
Query: 191 VALGAAVGQACLLLSAGTKGRRYMTPHA 218
V +G A +L+ G +R PHA
Sbjct: 158 VCMGLAASMGSFVLAGGKITQRLAFPHA 241
>TC18867 similar to UP|Q9ASV2 (Q9ASV2) At1g06730/F4H5_22, partial (25%)
Length = 748
Score = 34.7 bits (78), Expect = 0.022
Identities = 35/109 (32%), Positives = 50/109 (45%), Gaps = 17/109 (15%)
Frame = +2
Query: 59 SSPETLFN----------PSSSPDNLPFLDI---FDSPQLMATPAQLERSASYSQQRRPK 105
S PE L N PSS+P+ P ++ F S + TPA+ S S + RR +
Sbjct: 119 SPPECLSNLRFNSTPMDLPSSNPNPNPIQNLG*SFFSLSIAPTPAKASESPSLHRFRRRR 298
Query: 106 RPP--PDLPSLLLNGRIVYI--GMPLVPAVTELVIAELMFLQWMAPKEP 150
PP P+ P+LLL G V I M L + V L++ W+ + P
Sbjct: 299 IPPSEPNTPTLLLLGISVLILSSMSLNCLLLLSVNVRLIWSVWLPLRLP 445
>TC15031 weakly similar to UP|Q69107 (Q69107) LAT protein, partial (27%)
Length = 997
Score = 33.9 bits (76), Expect = 0.038
Identities = 34/127 (26%), Positives = 50/127 (38%), Gaps = 25/127 (19%)
Frame = +3
Query: 12 PTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLA-----SVAESSPETLFN 66
P+S P+SS+ F+P+ +S S NPN +SK + SV+ SS +
Sbjct: 492 PSSPPSSSSLFSPS----------VSSSTFVSNPNPSPISKASTASGPSVSSSSSSSSSG 641
Query: 67 PSSSPDNLPFLDIFDSPQLMATPAQLERSAS--------------------YSQQRRPKR 106
PS + P SP P E ++S S++RR K+
Sbjct: 642 PSPNSSGYPSSAAGTSPPSPIKPISAESTSS*IWASSNPRSSSRSSSSSTHRSRRRRRKK 821
Query: 107 PPPDLPS 113
P P L S
Sbjct: 822 PLPSLAS 842
>TC16031 similar to UP|O23548 (O23548) CLP proteinase homolog (CLP
proteinase like protein), partial (10%)
Length = 601
Score = 33.1 bits (74), Expect = 0.065
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = +2
Query: 257 AKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILW 293
AKH E S E + ++R Y A E+G+IDK+L+
Sbjct: 2 AKHIEKSPEQIEADIQRPKYFSPSEAVEYGIIDKVLY 112
>AV409919
Length = 423
Score = 32.7 bits (73), Expect = 0.085
Identities = 24/72 (33%), Positives = 28/72 (38%)
Frame = +3
Query: 42 PFNPNDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQ 101
P N + P SKL SP F+PSSSP S A+ + S
Sbjct: 111 PANASKPRTSKLPPPPLPSPTLYFSPSSSP---------SSTSSSTAGARRFGTPPLSTS 263
Query: 102 RRPKRPPPDLPS 113
P RPPP PS
Sbjct: 264 SPPPRPPPSSPS 299
>TC14473 similar to UP|Q8H1Y2 (Q8H1Y2) 20S proteasome alpha 6 subunit,
partial (86%)
Length = 1302
Score = 32.0 bits (71), Expect = 0.14
Identities = 29/105 (27%), Positives = 43/105 (40%)
Frame = +1
Query: 12 PTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSP 71
PTS +STR TP L T R + + + +P+ P A SSP T +PS S
Sbjct: 262 PTSSSPASTRLTPNSLLTRRRSSRLMTTSASPSPDSPL------TAVSSPATC-DPSVST 420
Query: 72 DNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLL 116
P P +S++ RP+ P D ++L+
Sbjct: 421 TLTPM-----------NPLSPLGGSSFTSPTRPRCAPSDHGNVLM 522
>BP038767
Length = 561
Score = 31.2 bits (69), Expect = 0.25
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 2/53 (3%)
Frame = +2
Query: 23 TPTRLRTLRTVN--AISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSPDN 73
TPTR N S S +P P P S +S + +PET NP +S DN
Sbjct: 266 TPTRRSEPAATNNKTPSPSPLPKKPPSPSPSTRSSAKKKTPETPTNPDASLDN 424
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 31.2 bits (69), Expect = 0.25
Identities = 34/122 (27%), Positives = 43/122 (34%), Gaps = 12/122 (9%)
Frame = +1
Query: 3 TCFRVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPE 62
TC P S PT P SS TP R+ T KSK + P AS A ++P
Sbjct: 265 TCLLAPASPPT--PKSSHSSTPPPSRSSSTT---PKSKNATSLTAPSCPPPASPAATAPA 429
Query: 63 TLFNPSSSPDNLPFLDIFDSPQLMATPAQLERS------------ASYSQQRRPKRPPPD 110
P + P N S TP+ L + +S Q +P PP
Sbjct: 430 PSKTPPNPPTNSSKPSSCKSSLPNTTPSSLSSAPPSPTTSPISTPSSPENQEKPASTPPS 609
Query: 111 LP 112
P
Sbjct: 610 AP 615
>TC9450 similar to GB|AAM16172.1|20334832|AY094016 At2g39720/T5I7.2
{Arabidopsis thaliana;}, partial (22%)
Length = 646
Score = 30.4 bits (67), Expect = 0.42
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Frame = +3
Query: 12 PTSMPASSTRFTPTRLRTLRTVNAISK-SKIPFNPNDPFLSKLASVAESSP 61
PT+ PAS R TPT R T+ ++ KIPF P + A+ E++P
Sbjct: 120 PTAKPASLKRSTPTNHRITLTLTVSARYLKIPFLPGG*VSAADAATPEAAP 272
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,782,137
Number of Sequences: 28460
Number of extensions: 60582
Number of successful extensions: 489
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 481
length of query: 322
length of database: 4,897,600
effective HSP length: 90
effective length of query: 232
effective length of database: 2,336,200
effective search space: 541998400
effective search space used: 541998400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC144541.11


