

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144539.10 - phase: 0
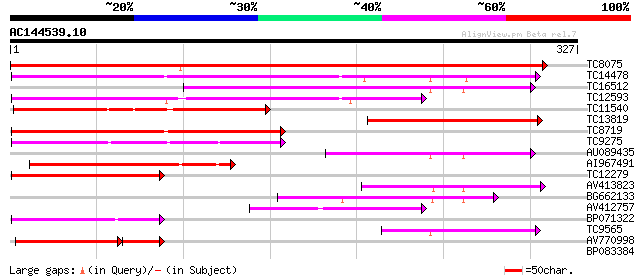
(327 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8075 similar to UP|E13B_PEA (Q03467) Glucan endo-1,3-beta-gluc... 479 e-136
TC14478 weakly similar to UP|Q94G86 (Q94G86) Beta-1,3-glucanase-... 203 4e-53
TC16512 similar to UP|Q9FXL4 (Q9FXL4) Elicitor inducible chitina... 154 2e-38
TC12593 146 5e-36
TC11540 similar to UP|AAR26001 (AAR26001) Endo-1,3-beta-glucanas... 129 5e-31
TC13819 similar to UP|Q9ZP12 (Q9ZP12) Glucan endo-1,3-beta-d-glu... 122 1e-28
TC8719 similar to UP|Q8H0I0 (Q8H0I0) Beta-1,3-glucanase-like pro... 101 2e-22
TC9275 similar to UP|Q9M4A9 (Q9M4A9) Beta-1,3 glucanase precurso... 90 6e-19
AU089435 85 1e-17
AI967491 82 1e-16
TC12279 weakly similar to UP|Q8H0I0 (Q8H0I0) Beta-1,3-glucanase-... 79 1e-15
AV413823 76 7e-15
BG662133 72 1e-13
AV412757 68 2e-12
BP071322 65 2e-11
TC9565 similar to UP|Q9MAQ2 (Q9MAQ2) CDS, partial (19%) 63 6e-11
AV770998 49 2e-09
BP083384 38 0.003
>TC8075 similar to UP|E13B_PEA (Q03467) Glucan endo-1,3-beta-glucosidase
precursor ((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) (Beta-1,3-endoglucanase) ,
partial (88%)
Length = 1437
Score = 479 bits (1234), Expect = e-136
Identities = 235/313 (75%), Positives = 270/313 (86%), Gaps = 3/313 (0%)
Frame = +2
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPS E + L KSNNI+RMRLYDPNQAAL+ALR+SGIEL+LGVPNSDLQ++ATN
Sbjct: 215 MMGNNLPSANEVVALYKSNNIRRMRLYDPNQAALQALRDSGIELILGVPNSDLQSLATNA 394
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQ--FAKFVLPAIQNIYQAIRAKNL 118
D A WVQ+NVLNF+PSV+IKYIAVGNEV+PVGG+ A++VLPA QNIYQAIRA+ L
Sbjct: 395 DNARNWVQRNVLNFWPSVRIKYIAVGNEVSPVGGAPTQWMAQYVLPATQNIYQAIRAQGL 574
Query: 119 HDQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKD 178
HDQIKV+TAID T+IG SYPPS+GSFRSDVRSYLDP IGYLVYA APL N+Y YFS+
Sbjct: 575 HDQIKVTTAIDTTLIGNSYPPSQGSFRSDVRSYLDPFIGYLVYAGAPLLVNVYPYFSHIG 754
Query: 179 NPKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPS 238
NP+D+SL YALFTSPN+V DG GYQNLFDA+LD++HAAIDNT IG+V+VVVSESGWPS
Sbjct: 755 NPRDVSLSYALFTSPNIVAQDGQYGYQNLFDAMLDAVHAAIDNTKIGYVEVVVSESGWPS 934
Query: 239 DGGFATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKNPELEKHFGVFYP 297
DGG AT+YDNAR+YLDNLIRHV GTP R + P ETYIF +FDENQK+PE EKHFG+F
Sbjct: 935 DGGSATSYDNARIYLDNLIRHVGRGTPRRPNKPTETYIFAMFDENQKSPEYEKHFGLFSA 1114
Query: 298 NKQKKYPFGFQGK 310
NKQKKYPFGF G+
Sbjct: 1115NKQKKYPFGFGGE 1153
>TC14478 weakly similar to UP|Q94G86 (Q94G86) Beta-1,3-glucanase-like
protein, partial (83%)
Length = 1784
Score = 203 bits (516), Expect = 4e-53
Identities = 113/314 (35%), Positives = 185/314 (57%), Gaps = 9/314 (2%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ +NLP + ++L +S +I ++RLY + ++AL NSGI +++G N D+ N+A + +
Sbjct: 166 VADNLPPPKSTVNLLRSTSIGKIRLYGADPEIIKALANSGIGIIIGAANGDIPNLAADPN 345
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
A+QWV NVL +YP+ I I VGNEV G A ++PAI+N+ A+ + +L +
Sbjct: 346 AAVQWVNSNVLPYYPASNITLITVGNEV-LTSGDPNLASQLVPAIRNVQSALTSASLGGK 522
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPK 181
IKVST M ++ S PPS GSF + + L ++ +L ++P N Y +F+Y+ +P+
Sbjct: 523 IKVSTVQSMAVLSQSDPPSSGSFNAAFQDNLKQLLQFLKDNHSPFTINPYPFFAYQSDPR 702
Query: 182 DISLQYALFTSPNVVVWDGSRG--YQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
+L + LF PN D G Y N+FDA +D++++A+ + G +++VV+E+GWPS
Sbjct: 703 PETLAFCLF-QPNKGRVDSGNGKLYTNMFDAQVDAVYSALSSLGFQGIEIVVAETGWPSR 879
Query: 240 GG---FATTYDNARVYLDNLIRHVKG--GTPMRSG-PIETYIFGLFDENQK-NPELEKHF 292
G + DNA+ Y NLI H++ GTP+ G ++TYIF ++DE+ K E+ F
Sbjct: 880 GDSNEVGPSVDNAKAYNGNLITHLRSLVGTPLMPGKSVDTYIFAIYDEDLKPGKGSERAF 1059
Query: 293 GVFYPNKQKKYPFG 306
G+F + Y G
Sbjct: 1060GLFKTDLTMSYDVG 1101
>TC16512 similar to UP|Q9FXL4 (Q9FXL4) Elicitor inducible chitinase
Nt-SubE76, partial (48%)
Length = 857
Score = 154 bits (389), Expect = 2e-38
Identities = 88/212 (41%), Positives = 128/212 (59%), Gaps = 9/212 (4%)
Frame = +2
Query: 101 FVLPAIQNIYQAIRAKNLHDQIKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYL 159
F++PA++NI+QA+ NLH IKVS+ I ++ +G SYP S GSFR + V+S P++ +L
Sbjct: 20 FLVPAMRNIHQALVKHNLHHDIKVSSPIALSALGNSYPSSSGSFRPELVQSVFKPMLEFL 199
Query: 160 VYANAPLFANIYSYFSYKDNPKDISLQYALF-TSPNVVVWDGSRGYQNLFDALLDSLHAA 218
+ L N+Y +F+Y+ N I L YALF +P V Y NLFDA +D++ AA
Sbjct: 200 RETGSYLMVNVYPFFAYESNADVIPLDYALFRQNPGVADPGNGLRYFNLFDAQIDAVFAA 379
Query: 219 IDNTGIGFVKVVVSESGWPSDGG---FATTYDNARVYLDNLIRHV--KGGTPMR-SGPIE 272
+ V +VVSE+GWPS G + +NA Y L+R + GTPMR +
Sbjct: 380 LSALKFNDVNIVVSETGWPSKGDENELGASAENAAAYNGGLVRKILTNSGTPMRPKADLT 559
Query: 273 TYIFGLFDENQK-NPELEKHFGVFYPNKQKKY 303
Y+F LF+ENQK P E++FG+FYP+++K Y
Sbjct: 560 VYLFALFNENQKPGPTSERNFGLFYPDQKKVY 655
>TC12593
Length = 846
Score = 146 bits (368), Expect = 5e-36
Identities = 89/246 (36%), Positives = 141/246 (57%), Gaps = 7/246 (2%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ +NLP L +S I ++RLY + A ++AL N+GI +++G N D+ +A++ +
Sbjct: 97 VADNLPPPSATAKLLQSTAIGKVRLYGTDPAIIKALANTGIGIVIGAANGDIPALASDPN 276
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEV---NPVGGSSQFAKFVLPAIQNIYQAIRAKNL 118
A WV NV +YP+ I I VGNEV N G +Q +LPAIQN+ A+ A +
Sbjct: 277 FAKTWVSSNVAPYYPASNIILITVGNEVITSNDTGLVNQ----MLPAIQNVQNALDAASF 444
Query: 119 -HDQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYK 177
+IKVST M ++G S PPS G F ++ + L ++ + +P N Y YF+YK
Sbjct: 445 GGGKIKVSTVHSMAVLGNSEPPSGGKFHAEYDTVLQGLLSFNNATGSPFAINPYPYFAYK 624
Query: 178 DNP-KDISLQYALFTSPNV--VVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSES 234
+P + +L + LF PN V + + Y N+FDA +D++ +A+D+ G V++VV+E+
Sbjct: 625 SDPGRPDNLAFCLF-QPNSGRVDPNTNLNYMNMFDAQVDAVRSALDSLGFKNVEIVVAET 801
Query: 235 GWPSDG 240
GWP G
Sbjct: 802 GWPYKG 819
>TC11540 similar to UP|AAR26001 (AAR26001) Endo-1,3-beta-glucanase, partial
(45%)
Length = 580
Score = 129 bits (325), Expect = 5e-31
Identities = 74/148 (50%), Positives = 95/148 (64%)
Frame = +3
Query: 3 GNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDI 62
G+NLPS++E +DL KS I RMR+YD ++ AL+ALR S IE++LGVPN LQ++ T+
Sbjct: 150 GDNLPSRQEVVDLYKSKGISRMRIYDQDEEALQALRGSNIEVILGVPNDKLQSL-TDAGA 326
Query: 63 AIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQI 122
A WV K V Y VKIKY+AVGNEV P A VLPA++NI AI + N QI
Sbjct: 327 AKDWVNKYV-KAYSDVKIKYMAVGNEVPP---GDAAAGSVLPAMRNIQNAISSAN*QGQI 494
Query: 123 KVSTAIDMTMIGTSYPPSKGSFRSDVRS 150
KVS AI +++ YPP G F + RS
Sbjct: 495 KVSLAIKTSLVANPYPPENGVFSDEARS 578
>TC13819 similar to UP|Q9ZP12 (Q9ZP12) Glucan endo-1,3-beta-d-glucosidase
precursor , partial (31%)
Length = 485
Score = 122 bits (305), Expect = 1e-28
Identities = 60/103 (58%), Positives = 76/103 (73%), Gaps = 2/103 (1%)
Frame = +3
Query: 207 LFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGGFATTYDNARVYLDNLIRHVKGGTPM 266
LFDA+LD ++AA++ G +KVVVSESGWPS GG A NA Y NLI+HVKGGTP
Sbjct: 3 LFDAILDGVYAALEKAGAPDMKVVVSESGWPSAGGDAANVQNAESYYKNLIQHVKGGTPK 182
Query: 267 R-SGPIETYIFGLFDENQK-NPELEKHFGVFYPNKQKKYPFGF 307
R +GPIETY+F +FDEN+K +PE E++FG+F P+K KY F
Sbjct: 183 RPNGPIETYLFAMFDENRKPDPETERNFGLFRPDKSAKYQINF 311
>TC8719 similar to UP|Q8H0I0 (Q8H0I0) Beta-1,3-glucanase-like protein,
partial (48%)
Length = 643
Score = 101 bits (251), Expect = 2e-22
Identities = 48/159 (30%), Positives = 96/159 (60%), Gaps = 1/159 (0%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ N+LP + ++L K+ + R++LYD + L AL NS I++++ +PN L + A +
Sbjct: 154 IANDLPQPEKVVELLKAQGLNRVKLYDTDATVLTALANSDIKVVVAMPNELLSSAAADQS 333
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQ 121
WV+ N+ +++P+ +I+ IAVGNEV KF++ A++N++ ++ + L
Sbjct: 334 FTDAWVKANISSYHPATQIEAIAVGNEV--FVDPKNTTKFLVAAMKNVHASLTKQGLDSA 507
Query: 122 IKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYL 159
IK+S+ + ++ + SYP S GSF+++ + + P++ +L
Sbjct: 508 IKISSPVALSALQNSYPASAGSFKTELIEPVIKPMLDFL 624
>TC9275 similar to UP|Q9M4A9 (Q9M4A9) Beta-1,3 glucanase precursor ,
partial (39%)
Length = 545
Score = 89.7 bits (221), Expect = 6e-19
Identities = 53/160 (33%), Positives = 92/160 (57%), Gaps = 2/160 (1%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNN-IKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
+G+NLP + K+ I R++++D N L+A +GI + + PN D+ + N
Sbjct: 73 LGDNLPPPAAVANFLKTRTTIDRVKIFDVNPQILQAFAGTGISVTVTAPNGDIAALR-NI 249
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWV ++ F+P +I YI VG+EV G + + ++PA++ +Y A++A+ + D
Sbjct: 250 DSARQWVVTHIKPFHPQTRINYILVGSEVLH-WGDTNMIRGLVPAMRTLYSALQAEGIRD 426
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSD-VRSYLDPIIGYL 159
IKV+TA + ++ +S PPS G FR + L P++ +L
Sbjct: 427 -IKVTTAHSLAIMRSSLPPSAGRFRPGFAKHVLGPMLKFL 543
>AU089435
Length = 423
Score = 85.1 bits (209), Expect = 1e-17
Identities = 54/129 (41%), Positives = 75/129 (57%), Gaps = 8/129 (6%)
Frame = +2
Query: 183 ISLQYALFT-SPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGG 241
IS+ YALF +P VV Y NLFDA LD++ AA+ VKV V+E+GW S G
Sbjct: 17 ISIDYALFKENPGVVDSGNGLKYTNLFDAQLDAVFAAMSALKYDDVKVAVTETGWLSSGD 196
Query: 242 ---FATTYDNARVYLDNLIRHV--KGGTPMR-SGPIETYIFGLFDENQK-NPELEKHFGV 294
+NA Y NL R V GTP+R ++ ++F LF+ENQK P E+++G+
Sbjct: 197 SNEVGAGQENAAAYNGNLARRVLSGAGTPLRPKEALDVFLFALFNENQKPGPTSERNYGL 376
Query: 295 FYPNKQKKY 303
FYP+++K Y
Sbjct: 377 FYPSEKKVY 403
>AI967491
Length = 385
Score = 82.0 bits (201), Expect = 1e-16
Identities = 43/119 (36%), Positives = 73/119 (61%)
Frame = +3
Query: 12 AIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIAIQWVQKNV 71
+++L KS KR+++YD N L+AL N+ I++ + VPN + NI+TN ++ W+Q NV
Sbjct: 27 SVNLIKSLKAKRVKIYDTNPEILKALENTDIQVSIMVPNELVTNISTNQTLSDHWIQSNV 206
Query: 72 LNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIKVSTAIDM 130
+ FY I+Y+ VGNE+ G+ + ++PA++ I ++ +LH +IKV T M
Sbjct: 207 VPFYRKTLIRYLLVGNELTSSTGNETW-PHIVPAMRRIKHSLTIFHLH-KIKVGTPFAM 377
>TC12279 weakly similar to UP|Q8H0I0 (Q8H0I0) Beta-1,3-glucanase-like
protein, partial (29%)
Length = 474
Score = 78.6 bits (192), Expect = 1e-15
Identities = 37/88 (42%), Positives = 57/88 (64%)
Frame = +1
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NNLPS + ++L KS + R+++YD + A L AL SGI++ + +PN L A
Sbjct: 190 IANNLPSATKVVNLLKSQGLNRVKVYDTDPAVLRALSGSGIKVTVDLPNQQLFAAARAPS 369
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEV 89
A+QWV++NV ++P +I+ IAVGNEV
Sbjct: 370 FALQWVERNVAAYHPHTQIEAIAVGNEV 453
>AV413823
Length = 396
Score = 76.3 bits (186), Expect = 7e-15
Identities = 44/113 (38%), Positives = 65/113 (56%), Gaps = 7/113 (6%)
Frame = +2
Query: 204 YQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGGFA---TTYDNARVYLDNLIRHV 260
Y N+FDA++D+ + A+ + + V+V+ESGWPS G + T DNA Y NLIRHV
Sbjct: 20 YTNVFDAIVDAAYFAMSDLNFTNIPVLVTESGWPSKGDSSEPDATIDNANTYNSNLIRHV 199
Query: 261 --KGGTPMRSG-PIETYIFGLFDEN-QKNPELEKHFGVFYPNKQKKYPFGFQG 309
GTP G + TYI+ L++E+ + P EK++G+F+ N Y G
Sbjct: 200 LNNTGTPKHPGIAVSTYIYELYNEDLRSGPVSEKNWGLFFANGGPVYTLHLTG 358
>BG662133
Length = 428
Score = 72.4 bits (176), Expect = 1e-13
Identities = 45/138 (32%), Positives = 71/138 (50%), Gaps = 10/138 (7%)
Frame = +3
Query: 155 IIGYLVYANAPLFANIYSYFSYKDNPKDISLQYALF----TSPNVVVWDGSRGYQNLFDA 210
++ +L N+ N Y Y+ Y ++YALF + +V + Y ++FDA
Sbjct: 15 LLQFLKNTNSSYMLNAYPYYGYTKGDGIFPIEYALFRPLPSVKQIVDPNTLYHYNSMFDA 194
Query: 211 LLDSLHAAIDNTGIGFVKVVVSESGWPSDGGF---ATTYDNARVYLDNLIRHV---KGGT 264
++D+ + +ID + +VV+E+GWPS GG T NA Y +N+IR V G
Sbjct: 195 MVDATYYSIDALNFKGIPIVVTETGWPSFGGANEPDATEKNAETYSNNMIRRVMNDSGPP 374
Query: 265 PMRSGPIETYIFGLFDEN 282
S PI TYI+ LF+E+
Sbjct: 375 SQPSIPINTYIYELFNED 428
>AV412757
Length = 315
Score = 68.2 bits (165), Expect = 2e-12
Identities = 37/103 (35%), Positives = 54/103 (51%), Gaps = 1/103 (0%)
Frame = +2
Query: 139 PSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFS-YKDNPKDISLQYALFTSPNVVV 197
PS G FR D+++ + I+ +L NAP NIY + S Y D + YA F +
Sbjct: 5 PSDGDFRPDIQNLMLQIVKFLSQNNAPFTVNIYPFISLYSD--ANFPGDYAFFNGFQSPI 178
Query: 198 WDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
D + Y N+ DA D+L A+ G G + ++V E GWP+DG
Sbjct: 179 TDNGKIYDNVLDANYDTLVWALQKNGFGNLPIIVGEIGWPTDG 307
>BP071322
Length = 428
Score = 64.7 bits (156), Expect = 2e-11
Identities = 35/88 (39%), Positives = 49/88 (54%)
Frame = +2
Query: 2 MGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNND 61
+ NNLP L S N+ R++LYD + L A NS +E ++G+ N DLQ++ +
Sbjct: 143 IANNLPPPSRVAVLINSLNVSRIKLYDADPNVLSAFANSNVEFIIGLGNQDLQSMRDPSK 322
Query: 62 IAIQWVQKNVLNFYPSVKIKYIAVGNEV 89
A WVQ+NV + KI I VGNEV
Sbjct: 323 -AQSWVQQNVQPYISQTKITCITVGNEV 403
>TC9565 similar to UP|Q9MAQ2 (Q9MAQ2) CDS, partial (19%)
Length = 872
Score = 63.2 bits (152), Expect = 6e-11
Identities = 37/98 (37%), Positives = 56/98 (56%), Gaps = 6/98 (6%)
Frame = +3
Query: 215 LHAAIDNTGIGFVKVVVSESGWPSDGG---FATTYDNARVYLDNLIR-HVKGGTPMR-SG 269
++AA+ G V V +SE+GWPS G T +NA+ Y N+I+ K GTP++
Sbjct: 15 VYAALSRLGCAKVPVHISETGWPSKGDDHQAGATVENAKRYNGNVIKISSKKGTPLKPDA 194
Query: 270 PIETYIFGLFDENQK-NPELEKHFGVFYPNKQKKYPFG 306
+ Y+F LF+ENQK P E+++G+F P+ Y G
Sbjct: 195 DLNIYVFALFNENQKPGPASERNYGLFKPDGTLAYSLG 308
>AV770998
Length = 507
Score = 48.9 bits (115), Expect(2) = 2e-09
Identities = 23/62 (37%), Positives = 39/62 (62%)
Frame = +3
Query: 4 NNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIA 63
+++P + + L K+ I+ +RLYD +QA L AL +GI++++ VPN +L I +N A
Sbjct: 126 SDMPHPTQVVALLKAQKIRNVRLYDADQAMLMALAKTGIQVVVTVPNEELLGIGQSNSTA 305
Query: 64 IQ 65
Q
Sbjct: 306 GQ 311
Score = 29.3 bits (64), Expect(2) = 2e-09
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +1
Query: 66 WVQKNVLNFYPSVKIKYIAVGNEV 89
WV +NV+ YP++ I I VG EV
Sbjct: 313 WVNRNVVAHYPAINITGIGVGAEV 384
>BP083384
Length = 362
Score = 37.7 bits (86), Expect = 0.003
Identities = 20/49 (40%), Positives = 29/49 (58%), Gaps = 2/49 (4%)
Frame = -1
Query: 263 GTPMR-SGPIETYIFGLFDENQK-NPELEKHFGVFYPNKQKKYPFGFQG 309
GTP R ++ YIF +F+ENQK P E+++G+F + Y GF G
Sbjct: 359 GTPHRPKNVMKAYIFAIFNENQKPGPFSERNYGLFKADGSISYNIGFHG 213
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,842,280
Number of Sequences: 28460
Number of extensions: 57535
Number of successful extensions: 309
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 285
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 285
length of query: 327
length of database: 4,897,600
effective HSP length: 91
effective length of query: 236
effective length of database: 2,307,740
effective search space: 544626640
effective search space used: 544626640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC144539.10


