

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144516.1 + phase: 0 /pseudo
(561 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
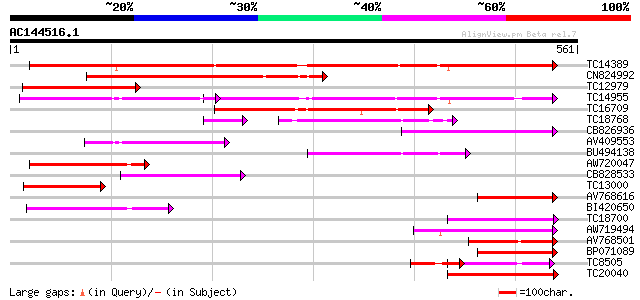
Score E
Sequences producing significant alignments: (bits) Value
TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 379 e-106
CN824992 316 6e-87
TC12979 similar to UP|Q9LQL2 (Q9LQL2) F5D14.23 protein, partial ... 194 4e-50
TC14955 UP|O22305 (O22305) Peptide transporter, complete 185 1e-47
TC16709 weakly similar to PIR|T47573|T47573 peptide transport-li... 178 2e-45
TC18768 118 3e-31
CB826936 102 2e-22
AV409553 100 5e-22
BU494138 98 4e-21
AW720047 98 4e-21
CB828533 90 1e-18
TC13000 similar to PIR|T47573|T47573 peptide transport-like prot... 89 1e-18
AV768616 89 1e-18
BI420650 86 2e-17
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 86 2e-17
AW719494 84 6e-17
AV768501 83 1e-16
BP071089 83 1e-16
TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partia... 82 2e-16
TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, ... 81 4e-16
>TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(72%)
Length = 1990
Score = 379 bits (973), Expect = e-106
Identities = 209/530 (39%), Positives = 326/530 (61%), Gaps = 7/530 (1%)
Frame = +3
Query: 20 LAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAVF 79
+A++GI NLVVFL + + ++NNVS W G V+ LVGA+++D+Y GRY T +
Sbjct: 45 MAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIA 224
Query: 80 QVIFVIGLMSLSLSSYLFLIKPKGC--GNESIQCGKHSSWEMGMFYLSIYLIALGNGGYQ 137
I+++G+ L+L+ L ++P C G E+ C + S W G+FYL++Y+IALG GG +
Sbjct: 225 SCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTK 404
Query: 138 PNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWL- 196
PNI+T GADQFDE KE K++FF+++ ++ G LFS T+LVY +D W+LG+ L
Sbjct: 405 PNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLP 584
Query: 197 SAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTK 256
+ G AF ++++FLVGTP YRH PSG+PL R QV VAA K K +P + L+ + +
Sbjct: 585 TVGLAF-SILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSME 761
Query: 257 ESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLP 316
E + + +I HT +FLD+AA+ + + ++PWRLC V+QVEE K + +++P
Sbjct: 762 EYANNSRNRIDHTSTLRFLDKAAVKTGK---------TSPWRLCTVTQVEETKQMTKMVP 914
Query: 317 IWLCTIIYSVVFTQMASLFVEQGDAMKTTV-YNFRVPPASMSCFDIISVAVFIFFYRRIL 375
I + T+I + Q +LF++QG + ++ NF +PPA +S IIS+ I Y R+
Sbjct: 915 ILITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVF 1094
Query: 376 DPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQDCRHCKGSS--- 432
P + + K + +G+T LQR+G GL++ V+ MV A + E RL+ A + +H G
Sbjct: 1095VPVIRRYTK-NPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVARE--KHLLGQHDII 1265
Query: 433 TLSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLV 492
LSIF IPQYAL G +E F + +++FF Q P+G+KS G + TS++LG ++SS L+
Sbjct: 1266PLSIFILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLL 1445
Query: 493 SVVMKISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWY 542
S V I+ + GWI NLN LD +Y + L+ ++ + ++ AK++
Sbjct: 1446STVADITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFF 1595
>CN824992
Length = 708
Score = 316 bits (810), Expect = 6e-87
Identities = 155/239 (64%), Positives = 192/239 (79%), Gaps = 1/239 (0%)
Frame = +2
Query: 77 AVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNGGY 136
A+FQVIFV+GL SLSL++YLFL++P GCG++ + C HSS++ +FY+SIYLIALGNGGY
Sbjct: 2 AIFQVIFVVGLASLSLTTYLFLLEPNGCGDKELPCTSHSSYQTALFYVSIYLIALGNGGY 181
Query: 137 QPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGFWL 196
QPNIATFGADQFDEE KE H+K++FFSYFYLALN G LFSNTIL YFED+G+W LGFW
Sbjct: 182 QPNIATFGADQFDEEDPKEQHSKISFFSYFYLALNLGSLFSNTILNYFEDDGLWTLGFWA 361
Query: 197 SAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEM-PSNGDDLYNMDT 255
SAG AF+AL+LFL GTP YR+FKP GNP+ RFCQV VAA RK KV++ P D L+ +
Sbjct: 362 SAGSAFLALVLFLCGTPWYRYFKPGGNPVPRFCQVFVAAMRKWKVKVSPGEEDQLH--EV 535
Query: 256 KESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRL 314
+E ST+ RK+ HT GF+FLD+AA+I+S++ Q+ CS W L V+QVEEVKCILRL
Sbjct: 536 EECSTNGGRKMFHTEGFRFLDKAALITSQE-SKQENECS-LWHLSTVTQVEEVKCILRL 706
>TC12979 similar to UP|Q9LQL2 (Q9LQL2) F5D14.23 protein, partial (25%)
Length = 601
Score = 194 bits (492), Expect = 4e-50
Identities = 92/117 (78%), Positives = 106/117 (89%)
Frame = +3
Query: 13 VNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGR 72
+NQ LATLAFFG+GVNLV+FLTRV+GQ+NA+AANNVSKWTGTVY+FSLVGAFLSDSYWGR
Sbjct: 249 LNQGLATLAFFGVGVNLVLFLTRVLGQDNADAANNVSKWTGTVYLFSLVGAFLSDSYWGR 428
Query: 73 YKTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLI 129
YKTCA+FQ IFV+GL+ LSLSSYL L++PKGCG+E + CGKHSS EMGMF YLI
Sbjct: 429 YKTCAIFQGIFVLGLVFLSLSSYLSLLRPKGCGSELLHCGKHSSLEMGMFSYQSYLI 599
>TC14955 UP|O22305 (O22305) Peptide transporter, complete
Length = 2300
Score = 185 bits (470), Expect = 1e-47
Identities = 116/356 (32%), Positives = 191/356 (53%), Gaps = 5/356 (1%)
Frame = +1
Query: 192 LGFWLSAGCAFIALILFLVGTPKYRH-FKPSGNPLARFCQVLVAASRKVKVEMPSNGDDL 250
L + LSA ++ I+F G+P YRH + + +P F +V + A R K+++P N +L
Sbjct: 667 LAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSEL 846
Query: 251 YNMDTKESSTDVNRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKC 310
+ + RKI HT F FLDRAAI S SNP C V+QVE K
Sbjct: 847 HEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTD------LSNP--PCTVTQVEGTKL 1002
Query: 311 ILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTVY-NFRVPPASMSCFDIISVAVFIF 369
+L + IWL +I + + +++FV QG M T+ FR+P AS+ CF +++ + +
Sbjct: 1003 VLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLP 1182
Query: 370 FYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQDCRHCK 429
Y PF+ + + + +G+ LQR+G+G+ I VIAM VE+ R+ + H
Sbjct: 1183 IYDHYFIPFMRR-RTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKK--HHIV 1353
Query: 430 GSST---LSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNY 486
G +SIFW +PQ ++G S F+ G LEFF Q+P+ +K G+ LC + ++ G+Y
Sbjct: 1354 GPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSY 1533
Query: 487 VSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWY 542
+++ LV+++ K++ WI NLN HLD +Y L +++++ ++ + Y
Sbjct: 1534 INTFLVTMIDKLN-------WIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGY 1680
Score = 119 bits (299), Expect = 1e-27
Identities = 64/199 (32%), Positives = 116/199 (58%)
Frame = +3
Query: 10 YGSVNQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSY 69
Y V + L A++G+G NLV F+T + ++ + + + W+G + ++GA+++D+Y
Sbjct: 129 YILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILGAYIADTY 308
Query: 70 WGRYKTCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLI 129
GR+ T +I+ IGL+ L L++ L ++P C N C + S+ ++ +FY S+Y I
Sbjct: 309 TGRFWTITFSLLIYAIGLVLLVLTTTLKSLRP-ACENGI--CREASNLQVALFYTSLYTI 479
Query: 130 ALGNGGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGM 189
A+G+G +PN++TFGADQFD+ +E KV+FF+++ G L + +VY +++
Sbjct: 480 AVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKN- 656
Query: 190 WALGFWLSAGCAFIALILF 208
GF L C + + L+
Sbjct: 657 -GXGFGLQFICYWFSTFLY 710
>TC16709 weakly similar to PIR|T47573|T47573 peptide transport-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(41%)
Length = 712
Score = 178 bits (452), Expect = 2e-45
Identities = 97/219 (44%), Positives = 141/219 (64%), Gaps = 2/219 (0%)
Frame = +1
Query: 203 IALILFLVGTPKYRHFKPSGNPLARFCQVLVAASRKVKVEMPSNGDDLYNMDTKESSTDV 262
+A++ F GT YR+ KP G+PL R CQV+VA+ RK V+ P + LY + ES+
Sbjct: 19 VAVVSFFSGTRLYRNQKPGGSPLTRMCQVVVASMRKCGVQAPDDKSLLYEIADTESAIKG 198
Query: 263 NRKILHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLPIWLCTI 322
+RK+ HT+ F D+AA++ +QK + +PWRLC +QVEE+K ILR+LP+W I
Sbjct: 199 SRKLDHTNELSFFDKAAVVV--QSSDQKDSV-DPWRLCTETQVEELKSILRMLPVWATGI 369
Query: 323 IYSVVFTQMASLFVEQGDAMKTTV--YNFRVPPASMSCFDIISVAVFIFFYRRILDPFVG 380
I++ V+ QM++LFV QG M T V NF++PPA++S FD +SV ++ Y I+ P
Sbjct: 370 IFATVYGQMSTLFVLQGQTMNTHVGNSNFKIPPAALSIFDTLSVIFWVPVYDWIIVPIAR 549
Query: 381 KLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLK 419
K GLT+LQRMG+GL I++ AMV+A I+E RL+
Sbjct: 550 KF-SGHKNGLTQLQRMGIGLFISIFAMVAAAILEVIRLR 663
>TC18768
Length = 738
Score = 118 bits (296), Expect(2) = 3e-31
Identities = 67/178 (37%), Positives = 103/178 (57%), Gaps = 1/178 (0%)
Frame = +2
Query: 267 LHTHGFKFLDRAAIISSRDHDNQKGTCSNPWRLCPVSQVEEVKCILRLLPIWLCTIIYSV 326
+HT F+FLD+A I R + T + WRLC V QVE+VK +L ++PI+ CTI+++
Sbjct: 233 VHTDKFRFLDKACI---RVEEAGSNTKKSSWRLCSVGQVEQVKILLSVIPIFSCTIVFNT 403
Query: 327 VFTQMASLFVEQGDAMKT-TVYNFRVPPASMSCFDIISVAVFIFFYRRILDPFVGKLKKS 385
+ Q+ + V+QG AM T +F +PPAS+ I + + + Y PF ++
Sbjct: 404 ILAQLQTFSVQQGSAMDTHLTKSFHIPPASLQSIPYILLIIVVPLYDTFFVPFARRITGH 583
Query: 386 DSKGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHADQDCRHCKGSSTLSIFWQIPQY 443
+S G++ L+R+G GL +A +MVSA ++E K D+ H K TLSIFW PQ+
Sbjct: 584 ES-GISPLRRIGFGLFLATFSMVSAALLEK---KRRDEALNHNK---TLSIFWITPQF 736
Score = 33.9 bits (76), Expect(2) = 3e-31
Identities = 19/44 (43%), Positives = 25/44 (56%)
Frame = +1
Query: 192 LGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFCQVLVAA 235
+GF +SA + LI + GT YR+ P G+ L QVLVAA
Sbjct: 28 VGFGISAAVMAMGLISLISGTFYYRNKPPQGSILTPIAQVLVAA 159
>CB826936
Length = 596
Score = 102 bits (254), Expect = 2e-22
Identities = 56/156 (35%), Positives = 90/156 (56%), Gaps = 1/156 (0%)
Frame = +3
Query: 388 KGLTELQRMGVGLVIAVIAMVSAGIVESYRLKHA-DQDCRHCKGSSTLSIFWQIPQYALI 446
+G+ LQR+G GLV+ +I MV+A + E RL A + + L+IF +PQ+AL
Sbjct: 30 RGIAMLQRLGAGLVMYIITMVTAWLSERKRLSVAREYNLLGQHDKIPLTIFILLPQFALT 209
Query: 447 GASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPG 506
G + F+ + L+FF Q P+G+KS G + TS +LG + SS L+S V ++ + G
Sbjct: 210 GVANNFVDIAMLDFFYDQAPEGMKSLGISYSTTSTALGCFFSSFLLSTVADLTKKHGHKG 389
Query: 507 WIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWY 542
W+ NLN HLD +Y +A L+ + + ++ AK +
Sbjct: 390 WVLDNLNVSHLDYYYAFMAILSVANFLCFLVAAKLF 497
>AV409553
Length = 433
Score = 100 bits (250), Expect = 5e-22
Identities = 51/143 (35%), Positives = 85/143 (58%)
Frame = +2
Query: 75 TCAVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNG 134
T + +++V+G++ L+++ L +KP C N C K S+ ++ FY ++Y A+G G
Sbjct: 2 TFTLSSIVYVMGMILLTVAVSLKSLKPT-CTNGI--CNKASTSQIVFFYTALYTTAIGAG 172
Query: 135 GYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLALNFGQLFSNTILVYFEDEGMWALGF 194
G +PNI+TFGADQFD+ + E K +FF+++ G L + LVY ++ W LG+
Sbjct: 173 GTKPNISTFGADQFDDFNPHEKQTKASFFNWWMFTSFLGALIATLGLVYIQENLGWGLGY 352
Query: 195 WLSAGCAFIALILFLVGTPKYRH 217
+ ++L++F VGTP YRH
Sbjct: 353 GIPTSGLLLSLVIFYVGTPMYRH 421
>BU494138
Length = 518
Score = 97.8 bits (242), Expect = 4e-21
Identities = 53/163 (32%), Positives = 89/163 (54%), Gaps = 1/163 (0%)
Frame = +2
Query: 295 NPWRLCPVSQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQGDAMKTTV-YNFRVPP 353
N R+ + QVEE+KC+ R+ PIW I+ Q + V Q M + F++P
Sbjct: 2 NSARVVSIQQVEEIKCLARIFPIWAAGILGFTAMAQQGTFTVSQAMKMDRHIGSKFQIPA 181
Query: 354 ASMSCFDIISVAVFIFFYRRILDPFVGKLKKSDSKGLTELQRMGVGLVIAVIAMVSAGIV 413
S+ I++ +++ FY R P V ++ K + G+T LQR+G+G+V +V++M+ AG+V
Sbjct: 182 GSLGVISFITIGLWVPFYDRFFVPAVRRITKHEG-GITLLQRIGIGMVFSVLSMIVAGLV 358
Query: 414 ESYRLKHADQDCRHCKGSSTLSIFWQIPQYALIGASEVFMYVG 456
E R A+ + + G + +S+ W PQ L+G E F +G
Sbjct: 359 EKVRRGVANSN-PNPLGIAPMSVMWLAPQLVLMGLCEAFNAIG 484
>AW720047
Length = 419
Score = 97.8 bits (242), Expect = 4e-21
Identities = 49/119 (41%), Positives = 72/119 (60%)
Frame = +3
Query: 20 LAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAVF 79
LA++GI NLV++LT + + AA NV+ + GT Y+ L+G+FL+D+YWGRY T VF
Sbjct: 51 LAYYGIATNLVIYLTTKLNEGTVSAAKNVNTFHGTCYLTPLIGSFLADAYWGRYWTITVF 230
Query: 80 QVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNGGYQP 138
+++IG+ L LS+ + +KP C N S F+L +Y IALG GG +P
Sbjct: 231 YAVYLIGMCILILSATIPALKPAECVNSVCTATLAQS---VXFFLGLYSIALGTGGIKP 398
>CB828533
Length = 557
Score = 89.7 bits (221), Expect = 1e-18
Identities = 44/124 (35%), Positives = 72/124 (57%)
Frame = +1
Query: 110 QCGKHSSWEMGMFYLSIYLIALGNGGYQPNIATFGADQFDEEHSKEGHNKVAFFSYFYLA 169
+C + S ++ + +++Y IA+G GG + +++ FG+DQFD +E V FF+ FY
Sbjct: 184 ECIEASGKQVALLLVALYTIAVGAGGIKSSVSGFGSDQFDTTDPREEKKMVFFFNRFYFF 363
Query: 170 LNFGQLFSNTILVYFEDEGMWALGFWLSAGCAFIALILFLVGTPKYRHFKPSGNPLARFC 229
++ G LFS +LVY +D GF + AG ++L L G P YR+ +P G+PL
Sbjct: 364 VSTGSLFSVLVLVYVQDNIGRGWGFGIPAGIMLVSLAFLLYGKPLYRYKRPQGSPLTLIW 543
Query: 230 QVLV 233
+VL+
Sbjct: 544 KVLI 555
>TC13000 similar to PIR|T47573|T47573 peptide transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(21%)
Length = 446
Score = 89.4 bits (220), Expect = 1e-18
Identities = 40/81 (49%), Positives = 59/81 (72%)
Frame = +3
Query: 14 NQCLATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRY 73
N+C LA++G+ NLV++ + + Q++A A+ NVS W+GT YI L+GAFL+DSY GRY
Sbjct: 201 NECCERLAYYGMSTNLVLYFKKQLNQHSATASKNVSNWSGTCYITPLIGAFLADSYLGRY 380
Query: 74 KTCAVFQVIFVIGLMSLSLSS 94
T A F +I+VIG+ L+LS+
Sbjct: 381 WTIACFSIIYVIGMTLLTLSA 443
>AV768616
Length = 396
Score = 89.4 bits (220), Expect = 1e-18
Identities = 42/79 (53%), Positives = 59/79 (74%)
Frame = -1
Query: 464 QTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFYFL 523
Q PD ++S SAL + ++SLG Y+SSLLV++V KIST++ GWIP NLN GH+D F++L
Sbjct: 393 QAPDAMRSLSSALSLLTVSLGQYLSSLLVTIVTKISTKNGGSGWIPDNLNYGHVDYFFWL 214
Query: 524 LAALTSMDLIAYIACAKWY 542
LA L+ ++LI Y+ AK Y
Sbjct: 213 LAVLSVLNLIVYLPVAKLY 157
>BI420650
Length = 576
Score = 85.9 bits (211), Expect = 2e-17
Identities = 50/146 (34%), Positives = 80/146 (54%)
Frame = +1
Query: 17 LATLAFFGIGVNLVVFLTRVVGQNNAEAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTC 76
L + F V+LV++ V+ + A +AN ++ + G+ ++ SLVG F+SD+Y R TC
Sbjct: 151 LDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVGGFISDTYLNRLTTC 330
Query: 77 AVFQVIFVIGLMSLSLSSYLFLIKPKGCGNESIQCGKHSSWEMGMFYLSIYLIALGNGGY 136
+F + V+ L+ L++ + L + P+ CG S G + M Y S+ L+ALG GG
Sbjct: 331 LLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIAV----MLYTSLGLLALGLGGV 498
Query: 137 QPNIATFGADQFDEEHSKEGHNKVAF 162
+ + FGADQFDE+ E F
Sbjct: 499 RGAMVAFGADQFDEKDPVEAKALATF 576
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 85.5 bits (210), Expect = 2e-17
Identities = 40/110 (36%), Positives = 65/110 (58%)
Frame = +1
Query: 434 LSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVS 493
LS +W + QY LIG +EVF VG LEF + PD +KS GSA + LG + ++++ S
Sbjct: 46 LSAYWLLIQYCLIGVAEVFCIVGLLEFLYEEAPDAMKSIGSAYAALAGGLGCFAATIINS 225
Query: 494 VVMKISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWYK 543
++ ++ ++ W+ N+N G D FY++L AL+ ++ +I A YK
Sbjct: 226 IIKSLTGKEGKESWLAQNINTGRFDYFYWILTALSLVNFCIFIYSAHRYK 375
>AW719494
Length = 473
Score = 84.0 bits (206), Expect = 6e-17
Identities = 42/145 (28%), Positives = 78/145 (52%), Gaps = 2/145 (1%)
Frame = +3
Query: 400 LVIAVIAMVSAGIVESYRLKHADQD--CRHCKGSSTLSIFWQIPQYALIGASEVFMYVGQ 457
L+ + + +V++ +E+ R K A + + G +S W PQ L G +E F +GQ
Sbjct: 3 LLFSFLHLVTSATLETIRRKRAIEAGYINNAHGVLNMSAMWLAPQLCLGGIAEAFNAIGQ 182
Query: 458 LEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHL 517
EF+ + P + S+L ++ GN VSS + S V I++ GW+ N+NKG
Sbjct: 183 NEFYYTEFPRTMSXVASSLLGLGMAAGNIVSSFVFSAVENITSSGGKEGWVSDNINKGRF 362
Query: 518 DRFYFLLAALTSMDLIAYIACAKWY 542
D++Y+++ ++ ++L+ Y+ C+ Y
Sbjct: 363 DKYYWVVXGISGLNLVYYLICSWAY 437
>AV768501
Length = 442
Score = 83.2 bits (204), Expect = 1e-16
Identities = 38/88 (43%), Positives = 62/88 (70%)
Frame = -1
Query: 455 VGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNK 514
+GQL+FF + P G+K+ L ++++SLG + SS+LV++V K++ GW+ NLN+
Sbjct: 442 IGQLDFFLRECPKGMKTMSPGLFLSTLSLGFFFSSMLVTLVHKMTGPRQ--GWLADNLNQ 269
Query: 515 GHLDRFYFLLAALTSMDLIAYIACAKWY 542
G LD FY+LLA L+ ++L+ ++ CAKWY
Sbjct: 268 GKLDDFYWLLAVLSGVNLVVFLFCAKWY 185
>BP071089
Length = 393
Score = 83.2 bits (204), Expect = 1e-16
Identities = 38/79 (48%), Positives = 56/79 (70%)
Frame = -3
Query: 464 QTPDGLKSFGSALCMTSISLGNYVSSLLVSVVMKISTEDHMPGWIPVNLNKGHLDRFYFL 523
+ PD ++S SAL +T+ +LGNYVS+LLV +V K++T GWIP N+N+GHLD FY+L
Sbjct: 385 EAPDAMRSLCSALSLTTNALGNYVSTLLVIIVTKVTTSSGSLGWIPDNMNRGHLDYFYWL 206
Query: 524 LAALTSMDLIAYIACAKWY 542
L L+ ++ + Y+ AK Y
Sbjct: 205 LTVLSLLNFLVYLWIAKRY 149
>TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partial (23%)
Length = 1495
Score = 82.4 bits (202), Expect = 2e-16
Identities = 46/106 (43%), Positives = 64/106 (59%)
Frame = +3
Query: 434 LSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVS 493
LS FW + F YVGQLEFF + P+ +KS + L + ++S+G +VSSLLVS
Sbjct: 216 LSFFWW-------EQGKAFAYVGQLEFFIREAPERMKSMSTGLFLAALSMGYFVSSLLVS 374
Query: 494 VVMKISTEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACA 539
+V K+S + W+ NLNKG LD FY+LLA L ++ I +I A
Sbjct: 375 IVDKLSKKK----WLKSNLNKGRLDYFYWLLAVLGVLNFILFIVLA 500
Score = 42.4 bits (98), Expect = 2e-04
Identities = 21/54 (38%), Positives = 33/54 (60%)
Frame = +2
Query: 397 GVGLVIAVIAMVSAGIVESYRLKHADQDCRHCKGSSTLSIFWQIPQYALIGASE 450
G+GLV + +AM+ + IVE R ++A K + +S FW +PQ+ L+GA E
Sbjct: 101 GIGLVFSFVAMMVSAIVEKERRENA------VKKHTNISAFWLVPQFFLVGAGE 244
>TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, partial
(20%)
Length = 578
Score = 81.3 bits (199), Expect = 4e-16
Identities = 38/111 (34%), Positives = 68/111 (61%), Gaps = 1/111 (0%)
Frame = +3
Query: 434 LSIFWQIPQYALIGASEVFMYVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLLVS 493
++ W QY +G++++F G +EFF + P ++S +AL S+S+G ++S++LVS
Sbjct: 3 MTFLWVAFQYLFLGSADLFTLAGMMEFFFTEAPWSMRSLATALSWASLSMGYFLSTVLVS 182
Query: 494 VVMKIS-TEDHMPGWIPVNLNKGHLDRFYFLLAALTSMDLIAYIACAKWYK 543
++ K++ H P + NLN HL+RFY+L+ L+ ++ I Y+ A YK
Sbjct: 183 IINKVTGAFGHTPWLLGSNLNHYHLERFYWLMCVLSGLNFIHYLFWANSYK 335
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,727,941
Number of Sequences: 28460
Number of extensions: 163008
Number of successful extensions: 1195
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 1150
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1165
length of query: 561
length of database: 4,897,600
effective HSP length: 95
effective length of query: 466
effective length of database: 2,193,900
effective search space: 1022357400
effective search space used: 1022357400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144516.1


