

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
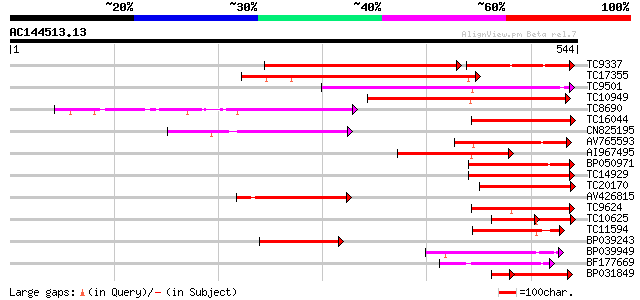
Query= AC144513.13 + phase: 0
(544 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B pre... 250 e-100
TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%) 249 6e-67
TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesteras... 231 3e-61
TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precu... 220 4e-58
TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precu... 168 2e-42
TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 137 4e-33
CN825195 127 5e-30
AV765593 125 2e-29
AI967495 124 4e-29
BP050971 117 5e-27
TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase ... 116 1e-26
TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isof... 113 7e-26
AV426815 110 8e-25
TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precur... 107 4e-24
TC10625 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 74 4e-22
TC11594 similar to UP|Q9FXW9 (Q9FXW9) Pectin methylesterase-like... 100 8e-22
BP039243 92 2e-19
BP039949 91 6e-19
BF177669 89 2e-18
BP031849 71 9e-18
>TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (54%)
Length = 1145
Score = 250 bits (638), Expect(2) = e-100
Identities = 121/189 (64%), Positives = 148/189 (78%)
Frame = +2
Query: 245 LDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVT 304
LDGSG Y T+ EAV AAP +S +R VIYVKKG+Y+E ++KKK NIMMVGDG+ T+++
Sbjct: 26 LDGSGNYTTVMEAVLAAPDYSMKRFVIYVKKGVYEEYAEIKKKKWNIMMVGDGMDVTVIS 205
Query: 305 SNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIE 364
NR+F GWTTF++ATFAVSG+GFIA+ +TF+NTAGP HQAVALR DSD + FFRCSI
Sbjct: 206 GNRSFGDGWTTFKSATFAVSGRGFIARGITFQNTAGPEKHQAVALRSDSDLAVFFRCSIT 385
Query: 365 GNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKS 424
G QD+LY H++RQFY+EC I GT+DFIFG+ AV QNC I R L QK TITAQGRK
Sbjct: 386 GYQDSLYPHTMRQFYKECRITGTVDFIFGDATAVFQNCNIQARKGLQDQKNTITAQGRKD 565
Query: 425 PHQSTGFTI 433
P QS+GF +
Sbjct: 566 PGQSSGFFV 592
Score = 131 bits (329), Expect(2) = e-100
Identities = 61/104 (58%), Positives = 76/104 (72%)
Frame = +3
Query: 439 LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSL 498
+ S TYLGRPWKEYSRTV++ ++MS +V +GWLEW +F DTL+YGEY NYG G+ +
Sbjct: 636 VGSIQTYLGRPWKEYSRTVFMQSFMSDVVSSQGWLEWNTSFD-DTLFYGEYLNYGAGAGV 812
Query: 499 AGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
RVKW GYHV+ D S AG FTV +F+ G WLP TGV +T GL
Sbjct: 813 GNRVKWGGYHVLND-SEAGKFTVNQFIEGNLWLPSTGVAYTGGL 941
>TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%)
Length = 848
Score = 249 bits (637), Expect = 6e-67
Identities = 124/244 (50%), Positives = 170/244 (68%), Gaps = 15/244 (6%)
Frame = +1
Query: 223 TEADQELLKSKPHGKIADAVVAL---DGSGQYRTINEAVNAAPSHSNRRH---VIYVKKG 276
T ++ L +S+ G + + V + +G+ Y +I +A+ AAP+H+ + +IYV++G
Sbjct: 94 TRTERILKESENQGVLLNDFVIVSPYNGTDNYTSIGDAIAAAPNHTKPQDGYFLIYVREG 273
Query: 277 LYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFR 336
Y+E + + K+ NI++VGDGI +TI+T N + + GWTTF ++TFAVSG+ F A D+TFR
Sbjct: 274 YYEEYVIVPKEKKNILLVGDGINKTIITGNHSVIDGWTTFNSSTFAVSGERFTAVDVTFR 453
Query: 337 NTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGA 396
NTAGP HQAVA+R ++D S F+RCS EG QDTLY HSLRQFYREC+IYGT+DFIFGN A
Sbjct: 454 NTAGPAKHQAVAVRNNADLSTFYRCSFEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAA 633
Query: 397 AVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYV---------LASQPTYLG 447
V QNC IY R PLP QK +TAQGR P+Q+TG +IQ+ + L S +YLG
Sbjct: 634 VVFQNCNIYARKPLPFQKNAVTAQGRTDPNQNTGISIQNCTIDAAPDLAAELNSTLSYLG 813
Query: 448 RPWK 451
RPW+
Sbjct: 814 RPWE 825
>TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase ,
partial (29%)
Length = 934
Score = 231 bits (588), Expect = 3e-61
Identities = 120/252 (47%), Positives = 150/252 (58%), Gaps = 9/252 (3%)
Frame = +2
Query: 300 QTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFF 359
+TI+T +NF G T +TATFA GFIAK +TF NTAGP HQAVALR DQSAF
Sbjct: 11 KTIITGRKNFADGVKTMQTATFANQAPGFIAKAITFENTAGPNGHQAVALRNQGDQSAFV 190
Query: 360 RCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITA 419
C I G QDTLY + RQFYR C I GTIDFIFG ++Q+ I R P Q T+TA
Sbjct: 191 GCHILGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSPTLIQHSIIVVRKPNDNQFNTVTA 370
Query: 420 QGRKSPHQSTGFTIQDSYVLASQ---------PTYLGRPWKEYSRTVYINTYMSSMVQPR 470
G + + TG IQD ++ +YLGRPWKEYSRTV + + + + P
Sbjct: 371 DGTPTKNMPTGIVIQDCEIVPEAALFPVRFKIKSYLGRPWKEYSRTVVMESTLGDFIHPE 550
Query: 471 GWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSW 530
GW W G+FALDTL+Y E+ N GPG++ AGR+KW GY + + A FT FL G
Sbjct: 551 GWFPWAGSFALDTLYYAEHANTGPGANTAGRIKWKGYRGVISRAEATQFTAGEFLKAG-- 724
Query: 531 LPRTGVKFTAGL 542
PR+G+ + L
Sbjct: 725 -PRSGLDWIKAL 757
>TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (34%)
Length = 773
Score = 220 bits (561), Expect = 4e-58
Identities = 107/201 (53%), Positives = 137/201 (67%), Gaps = 6/201 (2%)
Frame = +3
Query: 344 HQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCK 403
HQAVA+R SD+S F+RCS +G QDTLYAHS RQFYREC+I GTIDFIFGN A++ QNCK
Sbjct: 9 HQAVAMRSGSDRSVFYRCSFDGFQDTLYAHSNRQFYRECDITGTIDFIFGNAASIFQNCK 188
Query: 404 IYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA-----SQPTYLGRPWKEYSRTVY 458
I R P+P Q TITAQG+K P+Q+TG IQ S + + PTYLGRPWK++S T+
Sbjct: 189 IMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITPLESTYTAPTYLGRPWKDFSTTLI 368
Query: 459 INTYMSSMVQPRGWLEWLGNF-ALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAG 517
+ + + S ++P GW+ W+ N T++Y E++N GPGS +A RVKW GY A G
Sbjct: 369 MQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTADDLG 548
Query: 518 YFTVQRFLNGGSWLPRTGVKF 538
FT+ F+ G WLP T V F
Sbjct: 549 KFTLASFIQGPEWLPDTAVAF 611
>TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor
(Pectinesterase) , partial (59%)
Length = 1082
Score = 168 bits (426), Expect = 2e-42
Identities = 114/304 (37%), Positives = 170/304 (55%), Gaps = 14/304 (4%)
Frame = +3
Query: 44 CMDIENQNSCLTNI----HNELTRTGPPSPTSVINAALRTT--INEAIGAINNMTKISTF 97
C + +SCL ++ + ++ T P +I+ ++T I EA+ TK
Sbjct: 243 CEHALDASSCLAHVSEVSQSPISATKDPKLNILISLMTKSTSHIQEAMVK----TKAIKN 410
Query: 98 SVNN-REQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQD 156
+NN RE+ A+ DC++L+D S+ + S+ + + D T ++ + AWLS L+N
Sbjct: 411 RINNPREEAALSDCEQLMDLSIDRVWDSVMALTK----DNTDSHQ-DAHAWLSGVLTNHA 575
Query: 157 TCIEGFEGTDRRL-----ESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETS 211
TC++G EG R L E IS S T + L+S VL+ P+ +E
Sbjct: 576 TCLDGLEGPSRALMEAEIEDLISRSKTSLALLVS-VLA------------PKGG--NEQI 710
Query: 212 TDESLE--FPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRH 269
DE L+ FP W+T D+ LL+S A+ VVA DGSG+++T+ EAV +AP R+
Sbjct: 711 IDEPLDGDFPSWVTRKDRRLLESSVGDVNANVVVAKDGSGRFKTVAEAVASAPDSGKTRY 890
Query: 270 VIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFI 329
VIYVKKG YKENI++ KK TN+M+ GDG+ TI+T N N + G TTF++AT A G GFI
Sbjct: 891 VIYVKKGTYKENIEIGKKKTNVMLTGDGMDATIITGNLNVIDGSTTFKSATVAAVGDGFI 1070
Query: 330 AKDM 333
A+D+
Sbjct: 1071AQDI 1082
>TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (43%)
Length = 564
Score = 137 bits (345), Expect = 4e-33
Identities = 62/100 (62%), Positives = 73/100 (73%)
Frame = +3
Query: 444 TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVK 503
TYLGRPWK YSRTVY+ +Y+ V RGWLEW FALDTL+YGEY NYGPG+++ RV
Sbjct: 30 TYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVN 209
Query: 504 WPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
WPGY VI A A FTV +F+ G +WLP TGV F +GLS
Sbjct: 210 WPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGLS 329
>CN825195
Length = 551
Score = 127 bits (319), Expect = 5e-30
Identities = 67/184 (36%), Positives = 110/184 (59%), Gaps = 6/184 (3%)
Frame = +2
Query: 152 LSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYT----QLNRLPFRPPRNTTL 207
L+NQDTC +GF T ++ ++ ++ +++L+SN L+++ + +P + R +
Sbjct: 17 LTNQDTCADGFADTSGDVKDQMAVNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAM 196
Query: 208 HETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNR 267
+ + FP W+ D+ LL AD VV+ DG+G +TI EA+ P + +R
Sbjct: 197 RDDN------FPRWLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSR 358
Query: 268 RHVIYVKKGLYKE-NIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQG-WTTFRTATFAVSG 325
R +IYV+ G Y+E N+ + +K TN+M++GDG G+T++T +N M G TTF TA+FA SG
Sbjct: 359 RFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMYGLLTTFHTASFAASG 538
Query: 326 KGFI 329
GFI
Sbjct: 539 AGFI 550
>AV765593
Length = 501
Score = 125 bits (313), Expect = 2e-29
Identities = 62/122 (50%), Positives = 77/122 (62%), Gaps = 9/122 (7%)
Frame = -1
Query: 427 QSTGFTIQDSYVLASQP---------TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLG 477
Q TG +I + + A+ TYLGRPWKEYSRTVY+ T M +++ GW W G
Sbjct: 501 QDTGTSIHNCTIRAADDLASSNGATATYLGRPWKEYSRTVYMQTSMDNVIDSAGWRAWDG 322
Query: 478 NFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVK 537
FAL TL+Y E+ N GPGS+ GRV W GYHVI +A+ A FTV FL G WLP+TGV
Sbjct: 321 EFALSTLYYAEFNNSGPGSNTNGRVTWQGYHVI-NATDAANFTVSNFLLGDDWLPQTGVS 145
Query: 538 FT 539
+T
Sbjct: 144 YT 139
>AI967495
Length = 390
Score = 124 bits (311), Expect = 4e-29
Identities = 61/121 (50%), Positives = 79/121 (64%), Gaps = 10/121 (8%)
Frame = +1
Query: 373 HSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFT 432
H+ RQFY++C+IYGT+D IFGN A V QNCKI+ + PL Q ITAQGR P Q+TG T
Sbjct: 19 HAQRQFYKQCQIYGTVDLIFGNAAVVFQNCKIFAKKPLDGQANMITAQGRDDPFQNTGIT 198
Query: 433 IQDSYVLAS---------QPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLG-NFALD 482
I +S + A+ T LGRPW++YSR V + T+M ++V P GW W +FALD
Sbjct: 199 IHNSEIRAAPDLKPVVNKHNTSLGRPWQQYSRVVVLKTFMDTLVSPLGWSSWDDTDFALD 378
Query: 483 T 483
T
Sbjct: 379 T 381
>BP050971
Length = 477
Score = 117 bits (293), Expect = 5e-27
Identities = 55/103 (53%), Positives = 70/103 (67%), Gaps = 1/103 (0%)
Frame = -1
Query: 441 SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFAL-DTLWYGEYRNYGPGSSLA 499
S PT+LGRPWK +SRTV++ ++MS M+ P GWLEW D L++GEY N GPGS +A
Sbjct: 450 SIPTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETYTTYDDPLFFGEYPNKGPGSGVA 271
Query: 500 GRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
RVKW GYHV+ ++ A FTV F+ G WLP GV + AGL
Sbjct: 270 NRVKWRGYHVLNNSPAMD-FTVNPFIKGDLWLPSPGVSYHAGL 145
>TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase (Fragment)
, partial (24%)
Length = 693
Score = 116 bits (290), Expect = 1e-26
Identities = 55/102 (53%), Positives = 68/102 (65%)
Frame = +2
Query: 441 SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAG 500
S PTYLGRPWKEYSRTV++ + +S ++ P GW EW GNFAL+TL Y EY+ GPG+ +
Sbjct: 17 SFPTYLGRPWKEYSRTVFMQSSISDVIDPAGWHEWSGNFALNTLVYREYQYPGPGAGTSK 196
Query: 501 RVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
RV W G VI AS A FT F+ G +WL TG F+ GL
Sbjct: 197 RVAWKGVKVITSASEAQAFTPGSFIAGSNWLGSTGFPFSLGL 322
>TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (32%)
Length = 592
Score = 113 bits (283), Expect = 7e-26
Identities = 50/93 (53%), Positives = 64/93 (68%)
Frame = +1
Query: 451 KEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVI 510
K YSRTV++ +YM ++ P GWLEW FALDTL+YGEY+N GPGS+ + RVKW GY VI
Sbjct: 1 KLYSRTVFLKSYMEDLIDPAGWLEWNDTFALDTLYYGEYQNRGPGSNPSARVKWGGYRVI 180
Query: 511 KDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
++ A FTV +F+ G WL TG+ F LS
Sbjct: 181 NSSTEASQFTVGQFIQGSDWLNSTGIPFYFNLS 279
>AV426815
Length = 391
Score = 110 bits (274), Expect = 8e-25
Identities = 56/112 (50%), Positives = 79/112 (70%), Gaps = 1/112 (0%)
Frame = +3
Query: 218 FPEWMTEADQELLKSKPHGKI-ADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKG 276
+PEW++ AD+ LL++ G + AD VVA DGSG ++T++ AV AAP S R+VI +K G
Sbjct: 63 WPEWISAADRRLLQA---GTVKADVVVAADGSGNFKTVSAAVAAAPEKSKTRYVIKIKAG 233
Query: 277 LYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGF 328
+Y+EN+++ KK TNIM +GDG TI+T +RN + G TTF +AT AV G F
Sbjct: 234 VYRENVEVPKKKTNIMFLGDGRTTTIITGSRNVVDGSTTFHSATVAVVGGNF 389
>TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precursor
(Pectinesterase) (Pectin methylesterase) (Fragment) ,
partial (33%)
Length = 586
Score = 107 bits (268), Expect = 4e-24
Identities = 50/101 (49%), Positives = 62/101 (60%), Gaps = 2/101 (1%)
Frame = +3
Query: 444 TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFA--LDTLWYGEYRNYGPGSSLAGR 501
TYLGRPWK+YSRT+ + + + S + P GW EW L TL+YGEY N G G+ R
Sbjct: 18 TYLGRPWKKYSRTIVLQSSIDSHIDPTGWAEWDAQSKDFLQTLYYGEYLNSGAGAGTGKR 197
Query: 502 VKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
V WPG+HVIK A+ A FTV + + G WL GV F GL
Sbjct: 198 VNWPGFHVIKTAAEASKFTVAQLIQGNVWLKGKGVNFIEGL 320
>TC10625 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (32%)
Length = 538
Score = 73.6 bits (179), Expect(2) = 4e-22
Identities = 32/49 (65%), Positives = 38/49 (77%), Gaps = 3/49 (6%)
Frame = +2
Query: 463 MSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKW---PGYH 508
MS V PRGWLEW G+FALDTL+YGEY NYGPG+++ RVKW P Y+
Sbjct: 5 MSDHVHPRGWLEWNGDFALDTLYYGEYMNYGPGAAIGQRVKWTRVPDYY 151
Score = 48.1 bits (113), Expect(2) = 4e-22
Identities = 21/39 (53%), Positives = 26/39 (65%)
Frame = +3
Query: 505 PGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
PGY +I A FTV +F++G SWLP TGV + AGLS
Sbjct: 132 PGYRIITSTVEADRFTVAQFISGSSWLPSTGVAYLAGLS 248
>TC11594 similar to UP|Q9FXW9 (Q9FXW9) Pectin methylesterase-like protein,
partial (18%)
Length = 532
Score = 100 bits (248), Expect = 8e-22
Identities = 44/92 (47%), Positives = 61/92 (65%), Gaps = 4/92 (4%)
Frame = +3
Query: 445 YLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKW 504
YLGRPWKEYSRTV+I++ + ++V P+GW+ W G FAL TL+YGE+ N GPGS L+ RV W
Sbjct: 69 YLGRPWKEYSRTVFIHSLLEALVTPQGWMPWNGEFALKTLYYGEFENSGPGSDLSLRVSW 248
Query: 505 ----PGYHVIKDASAAGYFTVQRFLNGGSWLP 532
P HV+ ++ + F+ G W+P
Sbjct: 249 SSKVPAEHVLT-------YSAENFIQGDDWIP 323
>BP039243
Length = 462
Score = 92.4 bits (228), Expect = 2e-19
Identities = 43/81 (53%), Positives = 59/81 (72%)
Frame = -2
Query: 240 DAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIG 299
D VVA DG+G + I EAV AAP+ R VI++K G Y EN+++ +K TN+M VGDGIG
Sbjct: 443 DLVVAKDGTGNFAPIGEAVAAAPNSRATRFVIHIKAGAYFENVEVIRKKTNVMFVGDGIG 264
Query: 300 QTIVTSNRNFMQGWTTFRTAT 320
+T+V ++RN + GWTTF +AT
Sbjct: 263 KTVVKAHRNVVDGWTTFPSAT 201
>BP039949
Length = 517
Score = 90.5 bits (223), Expect = 6e-19
Identities = 48/134 (35%), Positives = 74/134 (54%), Gaps = 2/134 (1%)
Frame = -3
Query: 400 QNCKIYTRVPLPLQKVT--ITAQGRKSPHQSTGFTIQDSYVLASQPTYLGRPWKEYSRTV 457
+ C I++ L V+ ITAQGR+S ++ TGF+ + +L + +LGR W +S V
Sbjct: 509 EGCTIHSIAKPDLDGVSGSITAQGRQSINEETGFSFVNCTILGTGNVWLGRAWGAFSTAV 330
Query: 458 YINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAG 517
+ TYMS +V P GW +W T+++GEYR GPG++ RV + +KD A
Sbjct: 329 FSKTYMSEVVAPEGWNDWRDPSRDQTVFFGEYRCLGPGANYTSRVSYA--KQLKDYEANS 156
Query: 518 YFTVQRFLNGGSWL 531
Y + ++NG WL
Sbjct: 155 YMNIS-YINGNDWL 117
>BF177669
Length = 336
Score = 89.0 bits (219), Expect = 2e-18
Identities = 50/111 (45%), Positives = 64/111 (57%), Gaps = 1/111 (0%)
Frame = +3
Query: 413 QKVTIT-AQGRKSPHQSTGFTIQDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRG 471
QK+T+T QG + S SYV TYLGRPWK YS TV + + + S + P G
Sbjct: 15 QKLTLT*TQGISIQNCSISPFGNLSYV----QTYLGRPWKNYSTTVVMQSTLGSFISPNG 182
Query: 472 WLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQ 522
WL W+GN A DT++Y EY+N G G+S RVKW G I + A FTV+
Sbjct: 183 WLPWVGNSAPDTIFYAEYQNVGQGASTKNRVKWKGLKTI-TSKQASKFTVK 332
>BP031849
Length = 510
Score = 71.2 bits (173), Expect(2) = 9e-18
Identities = 30/60 (50%), Positives = 40/60 (66%)
Frame = +2
Query: 481 LDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTA 540
L + +YGEY N G G+S RV WPG+HV++ A A FTV RFL G W+P +GV F++
Sbjct: 71 LYSXYYGEYMNIGSGASTQNRVNWPGFHVLRSAGEAAPFTVSRFLQGDRWIPASGVPFSS 250
Score = 35.8 bits (81), Expect(2) = 9e-18
Identities = 13/22 (59%), Positives = 16/22 (72%)
Frame = +3
Query: 463 MSSMVQPRGWLEWLGNFALDTL 484
+ ++ PRGW EW GNFAL TL
Sbjct: 15 LDGLIHPRGWGEWRGNFALSTL 80
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,041,707
Number of Sequences: 28460
Number of extensions: 119782
Number of successful extensions: 632
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 609
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 610
length of query: 544
length of database: 4,897,600
effective HSP length: 95
effective length of query: 449
effective length of database: 2,193,900
effective search space: 985061100
effective search space used: 985061100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144513.13


