

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.8 - phase: 0
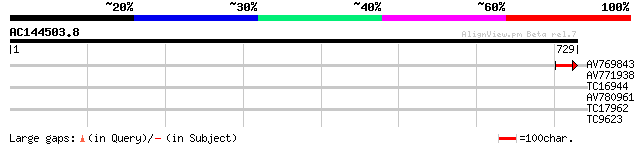
(729 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV769843 47 8e-06
AV771938 31 0.80
TC16944 similar to UP|Q94KL4 (Q94KL4) BEL1-like homeodomain 1, p... 31 0.80
AV780961 29 2.3
TC17962 UP|Q98GB0 (Q98GB0) Glucokinase, partial (29%) 28 4.0
TC9623 homologue to UP|Q8H1K8 (Q8H1K8) Stromal ascorbate peroxid... 27 8.9
>AV769843
Length = 365
Score = 47.4 bits (111), Expect = 8e-06
Identities = 24/28 (85%), Positives = 26/28 (92%)
Frame = -1
Query: 702 INGERELSSPTASVSAQSISSVRSHNSS 729
INGER+ SPTASVSAQSISSVRSH+SS
Sbjct: 365 INGERDPHSPTASVSAQSISSVRSHSSS 282
>AV771938
Length = 582
Score = 30.8 bits (68), Expect = 0.80
Identities = 20/65 (30%), Positives = 31/65 (46%)
Frame = +2
Query: 67 DSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
DS+ KE S L WL E + LDV+ +++ A + EG+ ++ LLLS
Sbjct: 260 DSKEKEASPLAAWLKEMKEGLDVV--RHKIQSLTAKVKEGQYPTADGFSYLEAKNLLLLS 433
Query: 127 LQSSI 131
S+
Sbjct: 434 YCQSL 448
>TC16944 similar to UP|Q94KL4 (Q94KL4) BEL1-like homeodomain 1, partial (29%)
Length = 1198
Score = 30.8 bits (68), Expect = 0.80
Identities = 17/66 (25%), Positives = 28/66 (41%)
Frame = -2
Query: 311 IQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDFFEDVGPLLSM 370
+ S++ M DW LT+PP + T + + F L + D D GPL +
Sbjct: 1107 VSPSSSCMKLNADWFLTFPPTGDVDGVDTETIGVFWFLATSWFELKLLDSVSDAGPLSCI 928
Query: 371 QLGGQA 376
+ G +
Sbjct: 927 FIAGDS 910
>AV780961
Length = 543
Score = 29.3 bits (64), Expect = 2.3
Identities = 14/36 (38%), Positives = 22/36 (60%)
Frame = -3
Query: 522 EWIPSLIFQSVVSWLLIFKNYRIRQQIDDLSIHIFL 557
E+ PS+ F S SW IFKN+ + ++D++ FL
Sbjct: 136 EYAPSICFFS--SWQAIFKNWCLASRLDEMVYECFL 35
>TC17962 UP|Q98GB0 (Q98GB0) Glucokinase, partial (29%)
Length = 435
Score = 28.5 bits (62), Expect = 4.0
Identities = 15/32 (46%), Positives = 17/32 (52%), Gaps = 2/32 (6%)
Frame = +1
Query: 453 KDDNRRRTTERQNRHPEQR--EWRRRLVGSVD 482
+D RRR Q+RH R WRRR GS D
Sbjct: 94 RDGARRRARPWQHRHVHHRYRHWRRRRPGSAD 189
>TC9623 homologue to UP|Q8H1K8 (Q8H1K8) Stromal ascorbate peroxidase,
partial (42%)
Length = 640
Score = 27.3 bits (59), Expect = 8.9
Identities = 15/46 (32%), Positives = 25/46 (53%)
Frame = -1
Query: 397 ESMEEEESFEDSGNKIVRMAETEAQQIALLANASLLADELLPRAAM 442
E EEEE E+ + E E++++AL+A ++ PRAA+
Sbjct: 190 EREEEEEEEENEKKEREEYEEEESERVALVAVEGIMRAAAPPRAAI 53
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.132 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,868,772
Number of Sequences: 28460
Number of extensions: 135831
Number of successful extensions: 745
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 744
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 745
length of query: 729
length of database: 4,897,600
effective HSP length: 97
effective length of query: 632
effective length of database: 2,136,980
effective search space: 1350571360
effective search space used: 1350571360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144503.8


