

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.3 + phase: 0
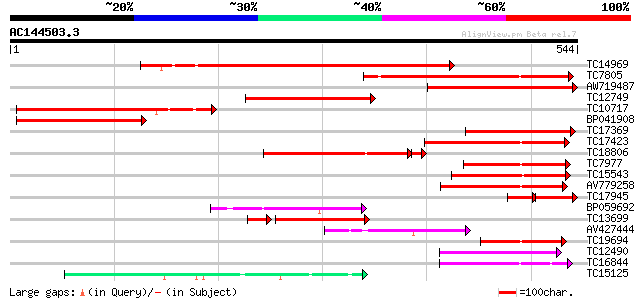
(544 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14969 similar to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (cl... 382 e-107
TC7805 homologue to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (c... 271 3e-73
AW719487 260 3e-70
TC12749 similar to UP|Q8W558 (Q8W558) 4-coumarate:CoA ligase , ... 224 3e-59
TC10717 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A lig... 216 8e-57
BP041908 196 6e-51
TC17369 similar to UP|Q8W558 (Q8W558) 4-coumarate:CoA ligase , ... 181 3e-46
TC17423 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like... 180 6e-46
TC18806 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A lig... 175 2e-45
TC7977 similar to UP|O48868 (O48868) 4-coumarate:CoA ligase 2, p... 150 5e-37
TC15543 similar to UP|Q84K86 (Q84K86) 4-coumarate:CoA ligase-lik... 134 5e-32
AV779258 113 7e-26
TC17945 homologue to UP|Q8W558 (Q8W558) 4-coumarate:CoA ligase ... 79 3e-22
BP059692 101 4e-22
TC13699 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like... 81 5e-19
AV427444 89 1e-18
TC19694 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-lik... 86 1e-17
TC12490 similar to UP|Q9FFE6 (Q9FFE6) AMP-binding protein (Adeno... 82 2e-16
TC16844 similar to UP|Q8LRT6 (Q8LRT6) Adenosine monophosphate bi... 78 4e-15
TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enz... 72 2e-13
>TC14969 similar to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (clone
GM4CL1B) - soybean (fragment) {Glycine
max;} , partial (70%)
Length = 928
Score = 382 bits (982), Expect = e-107
Identities = 188/313 (60%), Positives = 247/313 (78%), Gaps = 12/313 (3%)
Frame = +2
Query: 126 KLLVTQACYYDKVKDLENV------------KLVFVDSSPEEDNHMHFSELIQADQNEME 173
KL++TQA Y DK++ LE+V K+V VD P++ + FS + + ++++
Sbjct: 2 KLVITQAMYVDKLRPLEDVNGNDYPKLGEDFKVVTVDEPPKDC--LDFSVISEGKEDDLP 175
Query: 174 EVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCV 233
EV++N P+D VALP+SSGTTGLPKGV+LTHK L TS+ QQVDGENPNL +EDV+LCV
Sbjct: 176 EVEIN--PEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCV 349
Query: 234 LPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSP 293
LP+FHI+SLNSVLLC LRA + +LLMPKF+I + L+ K++V++A VVPP+VLA+AK+P
Sbjct: 350 LPLFHIFSLNSVLLCALRAGSGVLLMPKFEIGSLLELIQKHRVSVAMVVPPLVLALAKNP 529
Query: 294 ELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEP 353
++ ++DLSSIR++ SG APLGKELE+ +R++ P+A LGQGYGMTEAGPVL+M L FAK+P
Sbjct: 530 KVAEFDLSSIRLVLSGAAPLGKELEEALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQP 709
Query: 354 IDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDK 413
KSG+CGTVVRNAE+K++DP+ SL NQPGEICIRG QIMKGYLN+ +AT TID
Sbjct: 710 FPTKSGSCGTVVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDA 889
Query: 414 EGWLHTGDIGFID 426
EGWLHTGD+G+ID
Sbjct: 890 EGWLHTGDVGYID 928
>TC7805 homologue to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (clone
GM4CL1B) - soybean (fragment) {Glycine
max;} , partial (41%)
Length = 965
Score = 271 bits (692), Expect = 3e-73
Identities = 136/202 (67%), Positives = 162/202 (79%)
Frame = +3
Query: 340 GPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKG 399
GP + C S E I V+ G RNAE+K++DP+ SL NQPGEICIRG QIMKG
Sbjct: 15 GPKASFCWSIRAEDI-VRWGVWLGRQRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKG 191
Query: 400 YLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILS 459
YLN+ +AT TID EGWLHTGD+G+IDDDDE+FIVDR+KELIK+KGFQV PAELE +++S
Sbjct: 192 YLNDEKATATTIDAEGWLHTGDVGYIDDDDEIFIVDRVKELIKFKGFQVPPAELEGLLVS 371
Query: 460 HPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFID 519
HP I+D AVVP D AAGEVPVAFVVRSNG D TE+ +K+F+SKQVVFYKR+++V+F+
Sbjct: 372 HPSIADAAVVPQKDAAAGEVPVAFVVRSNG-FDLTEEAVKEFISKQVVFYKRLHKVYFVH 548
Query: 520 AIPKSPSGKILRKDLRAKLAAG 541
AIPKSPSGKILRKDLRAKL G
Sbjct: 549 AIPKSPSGKILRKDLRAKLENG 614
>AW719487
Length = 498
Score = 260 bits (665), Expect = 3e-70
Identities = 130/143 (90%), Positives = 137/143 (94%)
Frame = +2
Query: 402 NNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHP 461
N+PEAT TIDKEGWLHTGDIG+ID+DDELFIVDRLKELIKYKGFQVAPAELEA+ILSHP
Sbjct: 2 NDPEATERTIDKEGWLHTGDIGYIDEDDELFIVDRLKELIKYKGFQVAPAELEALILSHP 181
Query: 462 QISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAI 521
+ISDVAVVPM DEAAGEVPVAFVVRSNG DTTED+IK+FVSKQVVFYKRINRVFFIDAI
Sbjct: 182 KISDVAVVPMKDEAAGEVPVAFVVRSNGCTDTTEDEIKQFVSKQVVFYKRINRVFFIDAI 361
Query: 522 PKSPSGKILRKDLRAKLAAGVPN 544
PKSPSGKILRKDLRAKLAAGV N
Sbjct: 362 PKSPSGKILRKDLRAKLAAGVTN 430
>TC12749 similar to UP|Q8W558 (Q8W558) 4-coumarate:CoA ligase , partial
(23%)
Length = 380
Score = 224 bits (571), Expect = 3e-59
Identities = 111/125 (88%), Positives = 120/125 (95%)
Frame = +3
Query: 227 EDVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIV 286
EDVILCVLP+FHIYSLNSVLLCGLRAKA+ILLMPKFDI+A LV KY++TI PVVPPIV
Sbjct: 6 EDVILCVLPLFHIYSLNSVLLCGLRAKAAILLMPKFDIHALLPLVNKYRITIMPVVPPIV 185
Query: 287 LAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMC 346
LAIAKSP+L+K+DLSSIRVLKSGGAPLGKELED+VRAKFPKAKLGQGYGMTEAGPVLTMC
Sbjct: 186 LAIAKSPDLEKHDLSSIRVLKSGGAPLGKELEDSVRAKFPKAKLGQGYGMTEAGPVLTMC 365
Query: 347 LSFAK 351
LSFAK
Sbjct: 366 LSFAK 380
>TC10717 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
partial (32%)
Length = 691
Score = 216 bits (550), Expect = 8e-57
Identities = 105/195 (53%), Positives = 146/195 (74%), Gaps = 3/195 (1%)
Frame = +3
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
E++ IF+SKLPDI IP HLPLHSY FENL ++ RPCLI+ T +I+TY DV L ++K+A
Sbjct: 114 EEQFIFRSKLPDIPIPTHLPLHSYIFENLPRFQHRPCLIDGDTGKIHTYADVHLASRKIA 293
Query: 67 SGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
+GL+ LGI+ GDVIM++L NCP+F AFLGASF GA +T ANPF+T +E+AKQA A+ TK
Sbjct: 294 AGLHSLGIRHGDVIMLVLRNCPQFALAFLGASFAGAAVTTANPFYTQSELAKQATATKTK 473
Query: 127 LLVTQACYYDKVK---DLENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDD 183
L++TQ+ Y +K+ + ++ ++ +DS+ E +HFS L AD+N EV+ P+D
Sbjct: 474 LIITQSAYLEKINGFTESNDITVISIDST-ENRGALHFSVLTDADENRAPEVE--FSPND 644
Query: 184 VVALPYSSGTTGLPK 198
+VALP+SSGT+GLPK
Sbjct: 645 IVALPFSSGTSGLPK 689
>BP041908
Length = 487
Score = 196 bits (499), Expect = 6e-51
Identities = 94/126 (74%), Positives = 111/126 (87%), Gaps = 1/126 (0%)
Frame = +2
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
E++ IF+SKLPDIYIPKHLPLHSYCF+NL + SRPCLINAPT ++YTY+DV L A++ A
Sbjct: 104 EEQFIFRSKLPDIYIPKHLPLHSYCFQNLPDFASRPCLINAPTGDVYTYHDVDLAARRTA 283
Query: 67 SGLNKL-GIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNT 125
SGL+KL IQ+GDVIM+LLPN PEFVF+FL AS GA+ TAANPFFT+AEIAKQAKASNT
Sbjct: 284 SGLHKLCNIQKGDVIMILLPNSPEFVFSFLAASHLGAMATAANPFFTAAEIAKQAKASNT 463
Query: 126 KLLVTQ 131
KLL+TQ
Sbjct: 464 KLLITQ 481
>TC17369 similar to UP|Q8W558 (Q8W558) 4-coumarate:CoA ligase , partial
(20%)
Length = 580
Score = 181 bits (458), Expect = 3e-46
Identities = 91/106 (85%), Positives = 98/106 (91%)
Frame = +2
Query: 438 KELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDD 497
KELI YKGFQVAPA LEA++LSHPQISDVAVVPM DEAAGEVPVAFVVRSNG D ED+
Sbjct: 2 KELILYKGFQVAPAALEALLLSHPQISDVAVVPMKDEAAGEVPVAFVVRSNGHTDINEDE 181
Query: 498 IKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVP 543
IK+F+SKQVVFYKRINRVFFIDAIPKSPSGKILRK+LR+KL AGVP
Sbjct: 182 IKQFISKQVVFYKRINRVFFIDAIPKSPSGKILRKNLRSKLVAGVP 319
>TC17423 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like protein
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (25%)
Length = 655
Score = 180 bits (456), Expect = 6e-46
Identities = 83/139 (59%), Positives = 114/139 (81%)
Frame = +1
Query: 399 GYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIIL 458
GY NNPEAT+ T+DK+GW+HTGD+G+ D+D +L++VDR+KELIKYKGFQVAPAELE +++
Sbjct: 16 GYHNNPEATKLTLDKKGWVHTGDVGYFDEDGQLYVVDRIKELIKYKGFQVAPAELEGLLV 195
Query: 459 SHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFI 518
SHP+I D AVVP D+ AGEVP+A+VVRS S TE I++F++ QV +K++ RV FI
Sbjct: 196 SHPEILDAAVVPYPDDEAGEVPIAYVVRSPNS-SLTEKQIQEFIASQVAPFKKLRRVTFI 372
Query: 519 DAIPKSPSGKILRKDLRAK 537
++PK+ SGK+LR++L AK
Sbjct: 373 SSVPKTASGKLLRRELIAK 429
>TC18806 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
partial (26%)
Length = 529
Score = 175 bits (443), Expect(2) = 2e-45
Identities = 87/143 (60%), Positives = 111/143 (76%)
Frame = +1
Query: 244 SVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSI 303
S+LL +RA A +L++ KF+I ++ KY+VT+A VPPIVLA+ KS E +YDLSSI
Sbjct: 1 SILLSSMRAGAPVLILQKFEIVTLLEVIQKYRVTVASFVPPIVLALVKSGEAHQYDLSSI 180
Query: 304 RVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGT 363
RV+ +G AP+G ELE+ V+A+ P+AKLGQGYGMTEAGP L + L+FAKEP KSGACGT
Sbjct: 181 RVMITGAAPMGMELEEAVKARLPQAKLGQGYGMTEAGP-LAISLAFAKEPFKTKSGACGT 357
Query: 364 VVRNAEMKIVDPQNDSSLPRNQP 386
VVRNAEMKIVD + +SLP+ QP
Sbjct: 358 VVRNAEMKIVDTETGASLPKKQP 426
Score = 25.0 bits (53), Expect(2) = 2e-45
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +3
Query: 386 PGEICIRGDQIMKGY 400
PGEICIRG ++ K Y
Sbjct: 426 PGEICIRGTRL*KVY 470
>TC7977 similar to UP|O48868 (O48868) 4-coumarate:CoA ligase 2, partial
(18%)
Length = 638
Score = 150 bits (379), Expect = 5e-37
Identities = 76/103 (73%), Positives = 91/103 (87%)
Frame = +3
Query: 436 RLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTE 495
RLK LI KGFQVAPAELEA++++HP ISD AVVP+ DEAAGE+PVAF+VRSNGS T+
Sbjct: 3 RLKXLI*SKGFQVAPAELEALLIAHPNISDAAVVPLKDEAAGELPVAFIVRSNGS-KITD 179
Query: 496 DDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
D+IK+++SKQVVFYKRINRVFF DAIPK+ SGKILRK+L A+L
Sbjct: 180 DEIKQYISKQVVFYKRINRVFFTDAIPKAASGKILRKELTARL 308
>TC15543 similar to UP|Q84K86 (Q84K86) 4-coumarate:CoA ligase-like, partial
(21%)
Length = 579
Score = 134 bits (336), Expect = 5e-32
Identities = 66/114 (57%), Positives = 89/114 (77%)
Frame = +1
Query: 425 IDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFV 484
IDD++ +FIVDR+KELIKYKGFQVAPAELEAI+LSHP I D AVV + DE AGE+P A V
Sbjct: 1 IDDEENVFIVDRIKELIKYKGFQVAPAELEAILLSHPSIEDAAVVSLPDEEAGEIPAASV 180
Query: 485 VRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
V S G+ + +E+DI +V+ YK++ V F+++IPKSPSGKI+R+ ++ K+
Sbjct: 181 VLSLGAKE-SEEDIMNYVASNAASYKKVRVVHFVESIPKSPSGKIMRRLVKEKM 339
>AV779258
Length = 494
Score = 113 bits (283), Expect = 7e-26
Identities = 60/122 (49%), Positives = 85/122 (69%)
Frame = -2
Query: 414 EGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLD 473
+GWL TGD+ + D + ++IVDRLKELI+Y+G+QVAPAEL ++ SHP I DVAV+P D
Sbjct: 493 DGWLRTGDLCYFDHEGFMYIVDRLKELIQYQGYQVAPAELGHLLPSHPDIKDVAVIPXPD 314
Query: 474 EAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKD 533
AG VP+AFVVR + E +I +FV+ V ++I RV F+D+IPK+ + K +K
Sbjct: 313 ADAGPVPMAFVVRQPPT-SIGEAEIIEFVAHPVAP*QKIRRVAFVDSIPKNCNWKDYKKG 137
Query: 534 LR 535
L+
Sbjct: 136 LK 131
>TC17945 homologue to UP|Q8W558 (Q8W558) 4-coumarate:CoA ligase , partial
(11%)
Length = 526
Score = 78.6 bits (192), Expect(2) = 3e-22
Identities = 39/40 (97%), Positives = 39/40 (97%)
Frame = +3
Query: 505 QVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVPN 544
QVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGV N
Sbjct: 177 QVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVTN 296
Score = 43.5 bits (101), Expect(2) = 3e-22
Identities = 21/29 (72%), Positives = 23/29 (78%)
Frame = +1
Query: 478 EVPVAFVVRSNGSIDTTEDDIKKFVSKQV 506
E FVVRSNG DTTED+IK+FVSKQV
Sbjct: 1 EFGTRFVVRSNGCTDTTEDEIKQFVSKQV 87
>BP059692
Length = 486
Score = 101 bits (251), Expect = 4e-22
Identities = 57/153 (37%), Positives = 88/153 (57%), Gaps = 3/153 (1%)
Frame = -1
Query: 193 TTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRA 252
TTG KGV+ +H+ L+ V+ E +C +PMFHIY L +V GL
Sbjct: 486 TTGPSKGVISSHRNLIA----MVEIVRSRFSKEDERTFICTVPMFHIYGL-AVFATGLLI 322
Query: 253 KAS-ILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPEL--DKYDLSSIRVLKSG 309
S I+++ KF+++ F + K++ T P+VPPI++A+ + + KYDLSS+ + SG
Sbjct: 321 SGSTIVVLSKFEMHDMFSTIDKFRATFLPLVPPILVAMINNADAIKGKYDLSSLHTVLSG 142
Query: 310 GAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPV 342
GAPL KE+ + K+P + QGYG+TE+ V
Sbjct: 141 GAPLSKEVTEGFVDKYPNVTILQGYGLTESSGV 43
>TC13699 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like protein
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (23%)
Length = 391
Score = 80.9 bits (198), Expect(2) = 5e-19
Identities = 42/90 (46%), Positives = 57/90 (62%)
Frame = +3
Query: 256 ILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGK 315
++ M +F+ A V K+KVT VVPP+VLA+AK + KYDLSS++ + SG APLGK
Sbjct: 87 VVSMGRFEFEALLRAVEKHKVTNLWVVPPMVLALAKQSVVGKYDLSSLKYIGSGAAPLGK 266
Query: 316 ELEDTVRAKFPKAKLGQGYGMTEAGPVLTM 345
EL + K P + QGYGMTE V ++
Sbjct: 267 ELMEECARKLPHVSVCQGYGMTETCGVASL 356
Score = 30.4 bits (67), Expect(2) = 5e-19
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = +2
Query: 229 VILCVLPMFHIYSLNSVLLCGLR 251
V LCVLPMFH++ L + LR
Sbjct: 5 VFLCVLPMFHVFGLAIITYAALR 73
>AV427444
Length = 428
Score = 89.4 bits (220), Expect = 1e-18
Identities = 54/143 (37%), Positives = 78/143 (53%), Gaps = 3/143 (2%)
Frame = +1
Query: 303 IRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACG 362
+R++ SG +PL ++ + ++ F ++ +GYGMTE T C+ + D G G
Sbjct: 13 VRLMVSGASPLSPDIMEFLKICFG-GRVTEGYGMTE-----TTCVISCIDQGDKLGGHVG 174
Query: 363 TVVRNAEMKIVDPQNDSSLPRNQP---GEICIRGDQIMKGYLNNPEATRETIDKEGWLHT 419
+ E+K+VD + +QP GEIC+RG I KGY + TRE ID+EGWLHT
Sbjct: 175 SPSPACEVKLVDVPEMNYTSDDQPQPRGEICVRGPIIFKGYYKDEAQTREVIDEEGWLHT 354
Query: 420 GDIGFIDDDDELFIVDRLKELIK 442
GDIG L I+DR K + K
Sbjct: 355 GDIGTWLPGGRLKIIDRKKNIFK 423
>TC19694 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (16%)
Length = 504
Score = 86.3 bits (212), Expect = 1e-17
Identities = 44/83 (53%), Positives = 60/83 (72%)
Frame = +3
Query: 452 ELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKR 511
ELEA++L+H I D AV+P D+ AG+ P+A+VVR +G +E + FV++QV YKR
Sbjct: 3 ELEALLLTHSAILDAAVIPYPDKEAGQFPMAYVVRKDGG-SISEKQVMDFVAEQVAPYKR 179
Query: 512 INRVFFIDAIPKSPSGKILRKDL 534
I +V FI +IPK+ SGKILRKDL
Sbjct: 180 IRKVAFISSIPKNASGKILRKDL 248
>TC12490 similar to UP|Q9FFE6 (Q9FFE6) AMP-binding protein (Adenosine
monophosphate binding protein 5 AMPBP5), partial (21%)
Length = 387
Score = 82.4 bits (202), Expect = 2e-16
Identities = 44/118 (37%), Positives = 67/118 (56%), Gaps = 1/118 (0%)
Frame = +3
Query: 413 KEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPML 472
K GW +TGD+G + +D L I DR K++I G ++ E+E+++ HP ++ AVV
Sbjct: 27 KNGWFYTGDVGVMHEDGYLEIKDRSKDVIISGGENLSTVEVESVLYGHPAVNQAAVVARA 206
Query: 473 DEAAGEVPVAFVVRSNGSIDT-TEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKI 529
DE GE P AFV G +T TE ++ +F K + Y V F + +PK+ +GKI
Sbjct: 207 DEFWGETPCAFVSLKEGLKETVTEREVVEFCRKNMPHYMVPKTVVFKEELPKTSTGKI 380
>TC16844 similar to UP|Q8LRT6 (Q8LRT6) Adenosine monophosphate binding
protein 1 AMPBP1, partial (25%)
Length = 663
Score = 77.8 bits (190), Expect = 4e-15
Identities = 48/128 (37%), Positives = 66/128 (51%)
Frame = +3
Query: 413 KEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPML 472
K GW TGD+G D + + DR K++I G ++ ELE +I SHP + + AVV
Sbjct: 15 KGGWFRTGDLGVKHPDGYIELKDRSKDIIISGGENISSIELEGVIFSHPAVLEAAVVGRP 194
Query: 473 DEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRK 532
D GE P AFV G T +DI +F K++ Y V F D +PK+ +GK +
Sbjct: 195 DNYWGETPCAFVTLKEG-YSATAEDIIQFCHKRLPRYMAPRTVVFAD-LPKTSTGKTQKY 368
Query: 533 DLRAKLAA 540
LR K A
Sbjct: 369 VLREKAKA 392
>TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enzyme 12,
partial (48%)
Length = 1164
Score = 72.4 bits (176), Expect = 2e-13
Identities = 80/317 (25%), Positives = 121/317 (37%), Gaps = 26/317 (8%)
Frame = +1
Query: 53 YTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFT 112
+T+ +++A L L I + DV+ VL PN P GA++ N
Sbjct: 220 FTWSQTYERCRRLAFTLRSLNIAKNDVVSVLAPNIPAMYEMHFAVPMAGAVLNTINSRLD 399
Query: 113 SAEIAKQAKASNTKLLVTQACYYDKVKDLENVKLV---FVDSSPEEDNHMHFSELIQADQ 169
+ IA S K+ Y +K ++ + +V +S E+D ++ D
Sbjct: 400 AKNIATILSHSEAKIFFVDYEYVNKAREALRMLVVQKGVKPNSTEQDYSTLPLVIVIDDI 579
Query: 170 NEMEEVKV--------------NIKPDDV------VALPYSSGTTGLPKGVMLTHKGLVT 209
N V++ N P+++ +AL Y+SGTT PKGV+ +H+G
Sbjct: 580 NSPTGVRLGELEYEQMVHRGNPNYVPEEIQEEWSPIALNYTSGTTSEPKGVVYSHRGAYL 759
Query: 210 SIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASILL--MPKFDINAF 267
S V G SE V L LPMFH R ++ + DI
Sbjct: 760 STLSLVLGWEMG----SEPVYLWTLPMFHCNGWTFPWGVAARGGTNVCIRNCAASDIYRS 927
Query: 268 FGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPK 327
L + AP+V I+L P K + + +L G P LE F
Sbjct: 928 ISLHNVTHMCCAPIVFNIIL--GAKPSERKVIRNRVNILTGGAPPPASLLEQIEPLGF-- 1095
Query: 328 AKLGQGYGMTEA-GPVL 343
+ YG+TEA GP L
Sbjct: 1096-HVTHAYGLTEATGPAL 1143
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,207,902
Number of Sequences: 28460
Number of extensions: 102149
Number of successful extensions: 589
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 567
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 567
length of query: 544
length of database: 4,897,600
effective HSP length: 95
effective length of query: 449
effective length of database: 2,193,900
effective search space: 985061100
effective search space used: 985061100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144503.3


