

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144484.9 + phase: 0
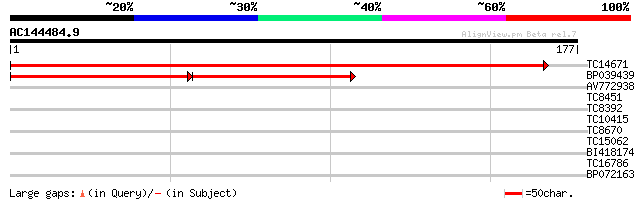
(177 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14671 similar to GB|AAM16192.1|20334872|AY094036 AT3g52300/T25... 302 2e-83
BP039439 101 6e-23
AV772938 28 0.90
TC8451 GB|CAA61589.1|897771|LJAS1GENE asparagine synthase (gluta... 28 1.2
TC8392 similar to UP|Q8LK52 (Q8LK52) Cp10-like protein, partial ... 27 2.0
TC10415 similar to UP|COM1_POPKI (Q43046) Caffeic acid 3-O-methy... 25 5.8
TC8670 weakly similar to UP|Q9ZRD1 (Q9ZRD1) NTGP4 (Fragment), pa... 25 7.6
TC15062 25 9.9
BI418174 25 9.9
TC16786 similar to GB|AAG26330.2|11072094|AF231996 MLL/GAS7 fusi... 25 9.9
BP072163 25 9.9
>TC14671 similar to GB|AAM16192.1|20334872|AY094036 AT3g52300/T25B15_70
{Arabidopsis thaliana;}, complete
Length = 988
Score = 302 bits (773), Expect = 2e-83
Identities = 143/168 (85%), Positives = 159/168 (94%)
Frame = +2
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPE 60
MSG KKV DVAFKAGK IDW+GMAKLLV+DEARREF NLRRAFD+VN+QL+TKFSQEPE
Sbjct: 59 MSGAGKKVVDVAFKAGKNIDWEGMAKLLVSDEARREFSNLRRAFDDVNSQLQTKFSQEPE 238
Query: 61 PIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKE 120
PIDW+YYRKGIG+RLVDMYK+HYESIE+PKFVDTVTP+YK KFD+LL+ELKEAEEKS KE
Sbjct: 239 PIDWDYYRKGIGSRLVDMYKEHYESIEVPKFVDTVTPKYKTKFDSLLIELKEAEEKSWKE 418
Query: 121 SERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
SERLEKEI +VQ LKK++STMTADEYFA+HPELKKKFDDEIRNDNWGY
Sbjct: 419 SERLEKEIADVQELKKKLSTMTADEYFAKHPELKKKFDDEIRNDNWGY 562
>BP039439
Length = 564
Score = 101 bits (252), Expect = 6e-23
Identities = 43/51 (84%), Positives = 50/51 (97%)
Frame = +1
Query: 58 EPEPIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLV 108
EPEPIDW+YYRKGIG+RLVDMYK+HYESIE+PKFVDTVTP+YK KFD+LL+
Sbjct: 412 EPEPIDWDYYRKGIGSRLVDMYKEHYESIEVPKFVDTVTPKYKTKFDSLLI 564
Score = 97.4 bits (241), Expect = 1e-21
Identities = 47/57 (82%), Positives = 52/57 (90%)
Frame = +3
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQ 57
MSG KKV DVAFKAGK IDW+GMAKLLV+DEARREF NLRRAFD+VN+QL+TKFSQ
Sbjct: 42 MSGAGKKVVDVAFKAGKNIDWEGMAKLLVSDEARREFSNLRRAFDDVNSQLQTKFSQ 212
>AV772938
Length = 441
Score = 28.1 bits (61), Expect = 0.90
Identities = 10/10 (100%), Positives = 10/10 (100%)
Frame = -3
Query: 158 DDEIRNDNWG 167
DDEIRNDNWG
Sbjct: 439 DDEIRNDNWG 410
>TC8451 GB|CAA61589.1|897771|LJAS1GENE asparagine synthase
(glutamine-hydrolysing) {Lotus corniculatus var.
japonicus;} , complete
Length = 2148
Score = 27.7 bits (60), Expect = 1.2
Identities = 18/57 (31%), Positives = 26/57 (45%)
Frame = +2
Query: 26 KLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQH 82
K++ DE R E + LRRAFD+ K + E + G+G +D K H
Sbjct: 1343 KMIKRDEGRIEKYILRRAFDDEEKPYLPKHILYRQK---EQFSDGVGYSWIDGLKDH 1504
>TC8392 similar to UP|Q8LK52 (Q8LK52) Cp10-like protein, partial (81%)
Length = 1014
Score = 26.9 bits (58), Expect = 2.0
Identities = 14/34 (41%), Positives = 22/34 (64%), Gaps = 3/34 (8%)
Frame = +1
Query: 88 IPKFVDTVTPQY---KPKFDALLVELKEAEEKSL 118
+ K V P++ KP D +LV++KEAEEK++
Sbjct: 253 VVKAATVVAPKHTAIKPLGDRVLVKIKEAEEKTV 354
>TC10415 similar to UP|COM1_POPKI (Q43046) Caffeic acid
3-O-methyltransferase 1
(S-adenosysl-L-methionine:caffeic acid
3-O-methyltransferase 1) (COMT-1) (CAOMT-1) , partial
(15%)
Length = 722
Score = 25.4 bits (54), Expect = 5.8
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = -2
Query: 149 EHPELKKKFDDEIRNDNWGYSH 170
+H +L++KFD I + WG H
Sbjct: 121 QHSKLQEKFD*SIVTEQWGKHH 56
>TC8670 weakly similar to UP|Q9ZRD1 (Q9ZRD1) NTGP4 (Fragment), partial
(71%)
Length = 1252
Score = 25.0 bits (53), Expect = 7.6
Identities = 21/75 (28%), Positives = 32/75 (42%), Gaps = 2/75 (2%)
Frame = +2
Query: 80 KQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEI--VNVQSLKKR 137
KQ Y+ ++ + + V + K L +LKE + LK E KEI + Q K
Sbjct: 797 KQEYDD-QLKRISEMVESKLKAATVRLEQQLKEEQAARLKAQENANKEIRELREQLFKAE 973
Query: 138 ISTMTADEYFAEHPE 152
+ A E +H E
Sbjct: 974 ENANKAREELRKHGE 1018
>TC15062
Length = 762
Score = 24.6 bits (52), Expect = 9.9
Identities = 9/27 (33%), Positives = 15/27 (55%)
Frame = +3
Query: 72 GTRLVDMYKQHYESIEIPKFVDTVTPQ 98
GTRL+ + H+ +I F+D P+
Sbjct: 234 GTRLISNFNYHHTVSDIRAFIDASRPE 314
>BI418174
Length = 602
Score = 24.6 bits (52), Expect = 9.9
Identities = 13/49 (26%), Positives = 24/49 (48%)
Frame = +2
Query: 91 FVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRIS 139
F+ P+YK D+ ++E + + + +E+E N+ KRIS
Sbjct: 266 FISDRFPKYKLGADSQILEETVEDNQGPSLKDVIEQEATNLSDQHKRIS 412
>TC16786 similar to GB|AAG26330.2|11072094|AF231996 MLL/GAS7 fusion protein
{Homo sapiens;} , partial (8%)
Length = 480
Score = 24.6 bits (52), Expect = 9.9
Identities = 10/17 (58%), Positives = 11/17 (63%)
Frame = -3
Query: 64 WEYYRKGIGTRLVDMYK 80
W YY K I TRLV +K
Sbjct: 340 WRYY*KSIKTRLVKAFK 290
>BP072163
Length = 396
Score = 24.6 bits (52), Expect = 9.9
Identities = 10/22 (45%), Positives = 14/22 (63%)
Frame = +3
Query: 81 QHYESIEIPKFVDTVTPQYKPK 102
Q Y S EIP+ + + PQ+ PK
Sbjct: 222 QPYHSHEIPQLIPFLLPQWLPK 287
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,291,297
Number of Sequences: 28460
Number of extensions: 21837
Number of successful extensions: 91
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 91
length of query: 177
length of database: 4,897,600
effective HSP length: 84
effective length of query: 93
effective length of database: 2,506,960
effective search space: 233147280
effective search space used: 233147280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC144484.9


