

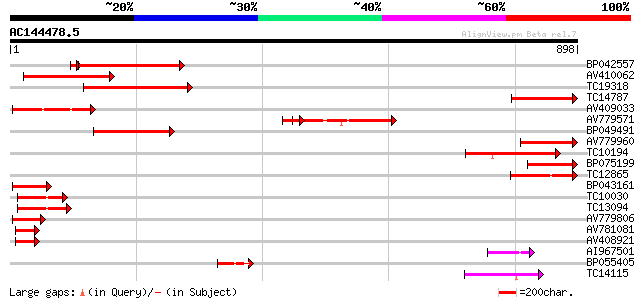
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144478.5 + phase: 0
(898 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP042557 314 7e-90
AV410062 290 5e-79
TC19318 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partia... 198 3e-51
TC14787 similar to UP|Q9ZTX8 (Q9ZTX8) ARF6 (Auxin response facto... 196 2e-50
AV409033 189 2e-48
AV779571 167 9e-42
BP049491 145 2e-35
AV779960 140 7e-34
TC10194 similar to UP|Q8S979 (Q8S979) Auxin response factor 7a (... 135 4e-32
BP075199 120 1e-27
TC12865 similar to UP|Q9ZTX8 (Q9ZTX8) ARF6 (Auxin response facto... 118 4e-27
BP043161 114 7e-26
TC10030 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partia... 100 8e-22
TC13094 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partia... 100 1e-21
AV779806 97 9e-21
AV781081 64 1e-10
AV408921 59 3e-09
AI967501 52 3e-07
BP055405 49 5e-06
TC14115 homologue to UP|O22465 (O22465) GH1 protein (Fragment), ... 47 1e-05
>BP042557
Length = 545
Score = 314 bits (805), Expect(2) = 7e-90
Identities = 156/168 (92%), Positives = 163/168 (96%), Gaps = 1/168 (0%)
Frame = +2
Query: 110 SKQPTN-YFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWK 168
SKQ TN YFCKTLTASDTSTHGGFSVPRRAAEKVFPPLD+SQQPPAQELIARDL+ NEWK
Sbjct: 41 SKQTTNNYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDYSQQPPAQELIARDLNDNEWK 220
Query: 169 FRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSS 228
FRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRA+R QT+MPSS
Sbjct: 221 FRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRANRSQTIMPSS 400
Query: 229 VLSSDSMHLGLLAAAAHAAATNSRFTIFYNPRACPSEFVIPLAKYVKA 276
VLSSDSMH+GLLAAAAHAA+TNSRFTIFYNPRA PSEFVIPLA+YVKA
Sbjct: 401 VLSSDSMHIGLLAAAAHAASTNSRFTIFYNPRASPSEFVIPLAQYVKA 544
Score = 34.7 bits (78), Expect(2) = 7e-90
Identities = 17/19 (89%), Positives = 17/19 (89%), Gaps = 1/19 (5%)
Frame = +3
Query: 97 QKEAYL-PAELGTPSKQPT 114
QKE YL PAELGTPSKQPT
Sbjct: 3 QKEVYLLPAELGTPSKQPT 59
>AV410062
Length = 435
Score = 290 bits (743), Expect = 5e-79
Identities = 140/144 (97%), Positives = 142/144 (98%)
Frame = +3
Query: 22 LVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHNLTMHADVETD 81
LVSLPA+GSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHN+TMHAD ETD
Sbjct: 3 LVSLPALGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHNVTMHADAETD 182
Query: 82 EVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTASDTSTHGGFSVPRRAAEK 141
EVYAQMTLQPLN QEQKEAYLPAELGTPSKQPTNYFCKTLTASDTSTHGGFSVPRRAAEK
Sbjct: 183 EVYAQMTLQPLNPQEQKEAYLPAELGTPSKQPTNYFCKTLTASDTSTHGGFSVPRRAAEK 362
Query: 142 VFPPLDFSQQPPAQELIARDLHGN 165
VFPPLDFSQQPPAQELIARDLHGN
Sbjct: 363 VFPPLDFSQQPPAQELIARDLHGN 434
>TC19318 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partial (23%)
Length = 585
Score = 198 bits (504), Expect = 3e-51
Identities = 98/172 (56%), Positives = 127/172 (72%)
Frame = +3
Query: 118 CKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQP 177
CKTLTASDTST GGFSV RR A++ PPLD S+QPP Q L+A+DLHG+EW+FRHIF GQP
Sbjct: 36 CKTLTASDTSTDGGFSVLRRHADECLPPLDMSKQPPTQALVAKDLHGSEWRFRHIFLGQP 215
Query: 178 KRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHL 237
+RHLL +GWSVFVS+KRLVAGD+ +F+ E +L +G+RRA R Q + SSV+SS S+HL
Sbjct: 216 RRHLLLSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVSSSVISSPSLHL 395
Query: 238 GLLAAAAHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFR 289
G+LA+A HA T S FT++Y PR PS ++ L + ++ MRF+
Sbjct: 396 GVLASAWHANLTGSMFTVYY*PRTSPSCNLLFLMINTWSPLPNNYTIAMRFK 551
>TC14787 similar to UP|Q9ZTX8 (Q9ZTX8) ARF6 (Auxin response factor 6),
partial (8%)
Length = 614
Score = 196 bits (497), Expect = 2e-50
Identities = 93/104 (89%), Positives = 98/104 (93%)
Frame = +2
Query: 795 LARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMG 854
LARMFGLEGELEDPVRSGWQLVFVDRE+DVLLLGDGPWPEFVNSVWCIKILSP+E+QQMG
Sbjct: 2 LARMFGLEGELEDPVRSGWQLVFVDRESDVLLLGDGPWPEFVNSVWCIKILSPQEMQQMG 181
Query: 855 NTGLGLLNSVPIQRLSNSICDDYVSRQDSRNLSSGITTVGSLDY 898
NTGL LLNSVP RLSN +CDDY+SRQD RNL SGITTVGSLDY
Sbjct: 182 NTGLELLNSVPNPRLSNGVCDDYMSRQDPRNLGSGITTVGSLDY 313
>AV409033
Length = 418
Score = 189 bits (479), Expect = 2e-48
Identities = 92/132 (69%), Positives = 110/132 (82%), Gaps = 1/132 (0%)
Frame = +3
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
GE++ ++SELWHACAGPLVSLP VGS VVYFPQGHSEQVA S KE D IP+YP+LP +
Sbjct: 27 GERKTINSELWHACAGPLVSLPPVGSLVVYFPQGHSEQVAASMQKETD-FIPSYPNLPSK 203
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELG-TPSKQPTNYFCKTLTA 123
LIC LHN+ +HAD ETDEVYAQMTLQP+N + K+A L +++G ++QPT +FCKTLTA
Sbjct: 204 LICMLHNVALHADPETDEVYAQMTLQPVNKYD-KDAILASDMGLKQNRQPTEFFCKTLTA 380
Query: 124 SDTSTHGGFSVP 135
SDTSTHGGFSVP
Sbjct: 381 SDTSTHGGFSVP 416
>AV779571
Length = 538
Score = 167 bits (422), Expect = 9e-42
Identities = 109/174 (62%), Positives = 122/174 (69%), Gaps = 10/174 (5%)
Frame = +1
Query: 449 LPGSLL--QFQQPPNFPNRTAAL-MQAQMLQQSQPQQAFQNNNQENQNLSQSQPQAQTNP 505
LP S+L F + +RT L +QAQM QQSQPQQ FQ NNQENQ L QS Q++
Sbjct: 40 LPNSILVPYFSFSKHRTSRTELL*VQAQM*QQSQPQQVFQ-NNQENQQLLQS----QSHL 204
Query: 506 QQHPQHQHSFNNQLHHHS-------QQQQQTQQQVVDNNQQISGSVSTMSQFVSATQPQS 558
QQH QHQHSFN Q +H+ QQQQQTQQQVVD NQQI + STMSQFVSA QPQS
Sbjct: 205 QQHLQHQHSFNGQTQNHNHNQQQQQQQQQQTQQQVVD-NQQIGNAGSTMSQFVSAPQPQS 381
Query: 559 PPPMQALSSLCHQQSFSDSNVNSSTTIVSPLHSIMGSSFPHDESSLLMSLPRTS 612
P QA SSL QQSFSDSNV TTIVSPLHS+MG SFP DE+S L++L RTS
Sbjct: 382 -SPTQAQSSLSQQQSFSDSNVKPVTTIVSPLHSLMG-SFPQDETSQLLNLSRTS 537
Score = 49.3 bits (116), Expect = 3e-06
Identities = 27/36 (75%), Positives = 28/36 (77%)
Frame = +3
Query: 432 AAAALQDMRTVVDPSKQLPGSLLQFQQPPNFPNRTA 467
AAAA MR+V DPSKQ P SLLQFQQ NFPNRTA
Sbjct: 3 AAAAHSGMRSV-DPSKQHPSSLLQFQQTQNFPNRTA 107
>BP049491
Length = 516
Score = 145 bits (367), Expect = 2e-35
Identities = 68/127 (53%), Positives = 92/127 (71%)
Frame = -3
Query: 134 VPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTGWSVFVSAK 193
VPRRAAE PPLD+ QQ P+QEL+A+DLHG EWKF HI+RG P+ HLLTTGWS+FVS
Sbjct: 514 VPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG*PRWHLLTTGWSIFVSQN 335
Query: 194 RLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAHAAATNSRF 253
LV+GD+V F+ E +L LGIRR+ P+ +P S + + + + +L+ A+A +T S F
Sbjct: 334 NLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSYPHILSVVANAISTRSMF 155
Query: 254 TIFYNPR 260
+FY+PR
Sbjct: 154 YVFYSPR 134
>AV779960
Length = 497
Score = 140 bits (354), Expect = 7e-34
Identities = 68/89 (76%), Positives = 74/89 (82%)
Frame = -3
Query: 810 RSGWQLVFVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMGNTGLGLLNSVPIQRL 869
RSGWQLVFVD+ENDVL+LGDGPWPEFVNSV CIKILSPEE+Q MG GLGL+ SVPIQRL
Sbjct: 495 RSGWQLVFVDQENDVLVLGDGPWPEFVNSVGCIKILSPEEMQPMGYHGLGLVYSVPIQRL 316
Query: 870 SNSICDDYVSRQDSRNLSSGITTVGSLDY 898
N DDY D RN SSG+TT+GSLDY
Sbjct: 315 CNGFSDDYTGPADPRNSSSGMTTLGSLDY 229
>TC10194 similar to UP|Q8S979 (Q8S979) Auxin response factor 7a (Fragment),
partial (13%)
Length = 1293
Score = 135 bits (339), Expect = 4e-32
Identities = 74/157 (47%), Positives = 95/157 (60%), Gaps = 7/157 (4%)
Frame = +1
Query: 723 SSSYMNTAGADSSLNHGVTPSIGESGFLHTQENGEQGNNP-------LNKTFVKVYKSGS 775
SS T G + + +I +S FL+ G Q P +T+ KVYK G+
Sbjct: 10 SSMVSQTFGVPDMTFNSIDSTIDDSSFLNGDPWGPQPAPPPPPAQFQRMRTYTKVYKRGA 189
Query: 776 FGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEF 835
GRS+DI ++S Y EL+ +LAR FG+EG+LED R GW+LV+VD E DVLL+GD PW EF
Sbjct: 190 VGRSIDIARYSGYEELKQDLARRFGIEGQLEDRQRIGWKLVYVDHEGDVLLVGDDPWEEF 369
Query: 836 VNSVWCIKILSPEEVQQMGNTGLGLLNSVPIQRLSNS 872
VN V CIKILSP+EVQQM G +P Q S+S
Sbjct: 370 VNCVRCIKILSPQEVQQMSLDGDFGSGGLPNQACSSS 480
>BP075199
Length = 426
Score = 120 bits (300), Expect = 1e-27
Identities = 56/78 (71%), Positives = 63/78 (79%)
Frame = -1
Query: 821 ENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMGNTGLGLLNSVPIQRLSNSICDDYVSR 880
ENDVL+LGDGPWP FVNS+ CIKILSPEE+QQMGN G GL+NSVPIQRL N DDY
Sbjct: 426 ENDVLVLGDGPWPGFVNSIGCIKILSPEEMQQMGNNGRGLVNSVPIQRLCNGFSDDYTGP 247
Query: 881 QDSRNLSSGITTVGSLDY 898
D N SSG+TT+GS+DY
Sbjct: 246 DDRSNSSSGMTTMGSVDY 193
>TC12865 similar to UP|Q9ZTX8 (Q9ZTX8) ARF6 (Auxin response factor 6),
partial (6%)
Length = 659
Score = 118 bits (296), Expect = 4e-27
Identities = 64/109 (58%), Positives = 79/109 (71%), Gaps = 4/109 (3%)
Frame = +2
Query: 794 ELARMFGLEGELEDPV--RSGWQLVFVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQ 851
ELAR+FG EG+LEDP+ RSGWQLVFVDREND+LLLGD PW EFVN+V IKILSP EV
Sbjct: 2 ELARIFGPEGQLEDPMTLRSGWQLVFVDRENDILLLGDDPWQEFVNNVSYIKILSPLEVP 181
Query: 852 QMGNTGLGLLNSVPIQRLSN--SICDDYVSRQDSRNLSSGITTVGSLDY 898
+MG + + S P +LSN + CDDY SR + R+ +G+ +GS Y
Sbjct: 182 EMGRS-VATSTSSPGHQLSNHGNSCDDYASRPELRSSRNGVAPMGSFHY 325
>BP043161
Length = 509
Score = 114 bits (285), Expect = 7e-26
Identities = 50/62 (80%), Positives = 57/62 (91%)
Frame = +2
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
GEK+ L+SELWHACAGPLVSLP G+RVVYFPQGHSEQV+ +TN+EVD H+PNYPSLPPQ
Sbjct: 323 GEKKCLNSELWHACAGPLVSLPTAGTRVVYFPQGHSEQVSATTNREVDGHMPNYPSLPPQ 502
Query: 65 LI 66
LI
Sbjct: 503 LI 508
>TC10030 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partial (13%)
Length = 588
Score = 100 bits (250), Expect = 8e-22
Identities = 45/79 (56%), Positives = 61/79 (76%)
Frame = +3
Query: 13 ELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHNL 72
ELWHACAGPLV++P G RV YFPQGH EQV STN+ + H+P Y LPP+++C++ N+
Sbjct: 354 ELWHACAGPLVTVPREGERVFYFPQGHIEQVEASTNQVAELHMPVY-DLPPKILCRVINV 530
Query: 73 TMHADVETDEVYAQMTLQP 91
+ A+ +TDEV+AQ+TL P
Sbjct: 531 MLKAEPDTDEVFAQVTLLP 587
>TC13094 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partial (18%)
Length = 685
Score = 100 bits (248), Expect = 1e-21
Identities = 46/85 (54%), Positives = 63/85 (74%)
Frame = +2
Query: 13 ELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHNL 72
ELWHACAGPLV++P G RV YFPQGH EQV STN+ D H+P Y LP +++C++ N+
Sbjct: 374 ELWHACAGPLVTVPREGERVFYFPQGHIEQVEASTNQVADEHMPVY-DLPSKILCRVINV 550
Query: 73 TMHADVETDEVYAQMTLQPLNAQEQ 97
+ A+ +TDEV+AQ+TL P Q++
Sbjct: 551 QLKAEPDTDEVFAQVTLLPEPNQDE 625
>AV779806
Length = 546
Score = 97.4 bits (241), Expect = 9e-21
Identities = 45/53 (84%), Positives = 49/53 (91%)
Frame = +2
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPN 57
GEK+ L+SEL HACAGPLVSLP +GSRVVYFPQGHSEQVA STN+EVDAHIPN
Sbjct: 386 GEKKCLNSELGHACAGPLVSLPPIGSRVVYFPQGHSEQVAASTNREVDAHIPN 544
>AV781081
Length = 528
Score = 63.9 bits (154), Expect = 1e-10
Identities = 28/38 (73%), Positives = 31/38 (80%)
Frame = +2
Query: 10 LDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVST 47
++ ELWHACAGPLVSLP +GS YFPQGH EQVA ST
Sbjct: 407 INPELWHACAGPLVSLPQLGSLAYYFPQGHIEQVAAST 520
>AV408921
Length = 429
Score = 59.3 bits (142), Expect = 3e-09
Identities = 26/37 (70%), Positives = 28/37 (75%)
Frame = +2
Query: 10 LDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVS 46
L ELWHACAGPLV+LP G RV YFPQGH EQ+ S
Sbjct: 317 LYKELWHACAGPLVTLPREGERVYYFPQGHMEQLEAS 427
>AI967501
Length = 393
Score = 52.4 bits (124), Expect = 3e-07
Identities = 26/75 (34%), Positives = 45/75 (59%), Gaps = 1/75 (1%)
Frame = +1
Query: 758 QGNNPLNKTFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLV 816
Q ++ KV G + GR++D+ Y++L EL ++F L+G+L+ R+ W++V
Sbjct: 169 QSKQICGRSRTKVQMQGVAVGRAVDLNTLDGYDQLIDELEQLFDLKGQLQH--RNKWEIV 342
Query: 817 FVDRENDVLLLGDGP 831
F D E D++L+GD P
Sbjct: 343 FTDDEGDMMLVGDDP 387
>BP055405
Length = 512
Score = 48.5 bits (114), Expect = 5e-06
Identities = 28/58 (48%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Frame = -3
Query: 330 DESTAGERQPRVSLWEIEPLTT-FPMYPSPFPLRLKRPWPPGLPSFHGMKDDDFGMSS 386
DEST GE RVS+W IEP+ T F YP P LR K PW PG+P DD++ + +
Sbjct: 405 DESTTGEHXSRVSIW*IEPVVTPFYGYP-PLFLRPKFPWXPGMP------DDEYDLEN 253
>TC14115 homologue to UP|O22465 (O22465) GH1 protein (Fragment), partial
(83%)
Length = 1610
Score = 47.0 bits (110), Expect = 1e-05
Identities = 40/149 (26%), Positives = 67/149 (44%), Gaps = 24/149 (16%)
Frame = +3
Query: 721 HQSSSYMNTAGADSSLNHGVTP--SIGESGFLHTQENGEQ--GNNPLNKTFVKVYKSGS- 775
H +S A A + G P S ++ T +N ++ G L+ FVKV G+
Sbjct: 699 HTGASISGNAPASKAQVVGWPPIRSFRKNSMATTSKNNDEVDGKPGLSALFVKVSMDGAP 878
Query: 776 FGRSLDITKFSSYNELRSELARMFGL------------------EGELEDPVR-SGWQLV 816
+ R +D+ +++Y EL S L +MF E +L D + S + L
Sbjct: 879 YLRKVDLRSYTTYQELSSALEKMFSCFTLGQCGSHGAPGRELLSESKLRDLLHGSEYVLS 1058
Query: 817 FVDRENDVLLLGDGPWPEFVNSVWCIKIL 845
+ D++ D +L+GD PW F + +KI+
Sbjct: 1059YEDKDGDWMLVGDVPWEMFTETCRRLKIM 1145
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.130 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,746,493
Number of Sequences: 28460
Number of extensions: 260495
Number of successful extensions: 2465
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 2170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2325
length of query: 898
length of database: 4,897,600
effective HSP length: 98
effective length of query: 800
effective length of database: 2,108,520
effective search space: 1686816000
effective search space used: 1686816000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144478.5


