

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
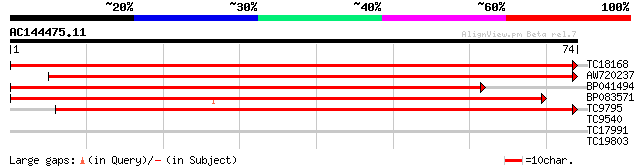
Query= AC144475.11 - phase: 0 /pseudo
(74 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat... 151 3e-38
AW720237 125 1e-30
BP041494 117 5e-28
BP083571 117 5e-28
TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment),... 106 7e-25
TC9540 26 1.6
TC17991 similar to UP|RIN4_ARATH (Q8GYN5) RPM1-interacting prote... 24 6.1
TC19803 similar to UP|O65568 (O65568) Serine carboxypeptidase II... 23 7.9
>TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (23%)
Length = 508
Score = 151 bits (381), Expect = 3e-38
Identities = 72/74 (97%), Positives = 73/74 (98%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 59 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 238
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 239 QVAMNPINTVFDAK 280
>AW720237
Length = 459
Score = 125 bits (315), Expect = 1e-30
Identities = 60/69 (86%), Positives = 63/69 (90%)
Frame = +3
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
+ AIGIDLGTTYSCVGVW +DRVEIIANDQGNRTTPSYV FT+SERLIGDAAKNQVA N
Sbjct: 33 QATAIGIDLGTTYSCVGVWMNDRVEIIANDQGNRTTPSYVGFTESERLIGDAAKNQVARN 212
Query: 66 PTNTVFDAK 74
P NTVFDAK
Sbjct: 213 PINTVFDAK 239
>BP041494
Length = 572
Score = 117 bits (292), Expect = 5e-28
Identities = 55/62 (88%), Positives = 58/62 (92%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
M G+GE PAIGIDLGTTYSCVGVWQ+DRVEII NDQGNRTTPSYVAFTD ERLIGDAAKN
Sbjct: 61 MVGEGESPAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDFERLIGDAAKN 240
Query: 61 QV 62
Q+
Sbjct: 241 QM 246
>BP083571
Length = 423
Score = 117 bits (292), Expect = 5e-28
Identities = 58/72 (80%), Positives = 63/72 (86%), Gaps = 2/72 (2%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
MA K EGPAIGIDLGTTYSCV VWQ +DR EII N+QGNRTTPS+VAFTD++RLIGDAA
Sbjct: 145 MATKYEGPAIGIDLGTTYSCVAVWQEQNDRAEIIHNEQGNRTTPSFVAFTDTQRLIGDAA 324
Query: 59 KNQVAMNPTNTV 70
KNQ A NPTNTV
Sbjct: 325 KNQAATNPTNTV 360
>TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment), partial
(63%)
Length = 981
Score = 106 bits (265), Expect = 7e-25
Identities = 50/68 (73%), Positives = 57/68 (83%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+VAF D ERLIG+AAKNQ A+NP
Sbjct: 145 GTVIGIDLGTTYSCVGVYRNGHVEIIANDQGNRITPSWVAFNDGERLIGEAAKNQFAVNP 324
Query: 67 TNTVFDAK 74
T+FD K
Sbjct: 325 ERTIFDVK 348
>TC9540
Length = 817
Score = 25.8 bits (55), Expect = 1.6
Identities = 13/34 (38%), Positives = 16/34 (46%)
Frame = -3
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTP 42
+IG+ VG WQ+D IA Q N TP
Sbjct: 308 SIGVATKLASITVGAWQYDPSNAIAIAQRNTVTP 207
>TC17991 similar to UP|RIN4_ARATH (Q8GYN5) RPM1-interacting protein 4,
partial (14%)
Length = 598
Score = 23.9 bits (50), Expect = 6.1
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = +3
Query: 15 GTTYSCVGVWQ 25
GTT+SC VWQ
Sbjct: 132 GTTFSCTKVWQ 164
>TC19803 similar to UP|O65568 (O65568) Serine carboxypeptidase II - like
protein (Serine carboxypeptidase II-like protein),
partial (36%)
Length = 593
Score = 23.5 bits (49), Expect = 7.9
Identities = 12/43 (27%), Positives = 20/43 (45%)
Frame = -2
Query: 28 RVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNTV 70
R+ I ++ +PS+V D R++ A + PTN V
Sbjct: 262 RILICLHEPVEECSPSFVIDRDVARIVCKAGVEVLTREPTNLV 134
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 965,472
Number of Sequences: 28460
Number of extensions: 8054
Number of successful extensions: 33
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32
length of query: 74
length of database: 4,897,600
effective HSP length: 50
effective length of query: 24
effective length of database: 3,474,600
effective search space: 83390400
effective search space used: 83390400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 48 (23.1 bits)
Medicago: description of AC144475.11


