

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.15 - phase: 0 /pseudo
(139 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
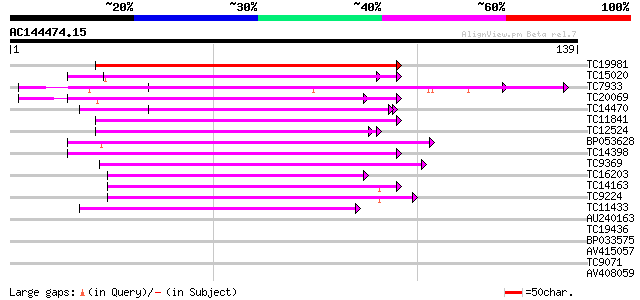
Score E
Sequences producing significant alignments: (bits) Value
TC19981 weakly similar to UP|AAP20228 (AAP20228) Resistance prot... 77 8e-16
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 50 1e-07
TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonas... 48 7e-07
TC20069 similar to UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kina... 45 5e-06
TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhi... 44 8e-06
TC11841 weakly similar to GB|BAC57345.1|28411900|AP005473 recept... 44 1e-05
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 43 2e-05
BP053628 42 5e-05
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 41 9e-05
TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resista... 40 2e-04
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 39 4e-04
TC14163 39 4e-04
TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Ara... 38 6e-04
TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor fa... 38 6e-04
AU240163 37 0.001
TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-li... 37 0.002
BP033575 37 0.002
AV415057 35 0.004
TC9071 similar to UP|Q9LT85 (Q9LT85) Similarity to receptor prot... 34 0.011
AV408059 34 0.011
>TC19981 weakly similar to UP|AAP20228 (AAP20228) Resistance protein SlVe1
precursor, partial (9%)
Length = 403
Score = 77.4 bits (189), Expect = 8e-16
Identities = 39/75 (52%), Positives = 52/75 (69%)
Frame = +1
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L L +S +L GP+DSSL L++L+++ L NN SS VP +FANF NLTTL+L C
Sbjct: 16 LKRLVILDLSSNNLFGPIDSSLADLKSLSILQLSHNNLSSTVPDSFANFSNLTTLHLINC 195
Query: 82 GLIGTFPQNIFQIKS 96
L G FP+ IF IK+
Sbjct: 196SLNGYFPKEIFHIKT 240
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 50.4 bits (119), Expect = 1e-07
Identities = 23/73 (31%), Positives = 41/73 (55%)
Frame = +1
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
++ ++++ SG L +++L L + L EN+FS P+P + ++ NL TL LR
Sbjct: 232 NINQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSF 411
Query: 84 IGTFPQNIFQIKS 96
G+ P +I +KS
Sbjct: 412 SGSIPPSITTLKS 450
Score = 47.8 bits (112), Expect = 7e-07
Identities = 26/81 (32%), Positives = 43/81 (52%), Gaps = 4/81 (4%)
Frame = +1
Query: 15 WSNALLPL----RDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANF 70
+S +L PL L L +++ SGP+ SSL+ L NL + L N+FS +P +
Sbjct: 265 YSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSGSIPPSITTL 444
Query: 71 KNLTTLNLRKCGLIGTFPQNI 91
K+L +L+ L G+ P ++
Sbjct: 445 KSLYSLDFSHNSLSGSLPNSM 507
>TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonase
inhibiting protein, partial (78%)
Length = 1320
Score = 47.8 bits (112), Expect = 7e-07
Identities = 45/149 (30%), Positives = 71/149 (47%), Gaps = 14/149 (9%)
Frame = +3
Query: 3 LDGISITSRGHEWSNA-------LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLG 55
L +SIT W+N L L++L L +S +LSG + +SL+RL NL + L
Sbjct: 372 LKSLSIT-----WTNVSGPIPDFLAQLKNLDSLYLSFNNLSGHIPASLSRLPNLLSLNLD 536
Query: 56 ENNFSSPVPQTFANFKNL-TTLNLRKCGLIGTFPQNIFQIKSQLLTY----LTTPTSMVF 110
N + P+P +F FK L L L G P ++ Q+ + + + LT S++F
Sbjct: 537 RNKLTGPIPASFGFFKKPGPNLFLSHNQLSGPIPSSLGQLDPERIDFSRNKLTGNASLLF 716
Query: 111 F--QTTHLVNLFIALYCVTLSSLEHVHTL 137
+ T L++L L LS +E +L
Sbjct: 717 AHNKKTQLLDLSRNLLSFDLSKVEFPKSL 803
Score = 37.7 bits (86), Expect = 7e-04
Identities = 27/95 (28%), Positives = 44/95 (45%), Gaps = 7/95 (7%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L+GP+ ++ +L L + + N S P+P A KNL +L L L G P ++ ++
Sbjct: 330 LTGPIQPAIAKLTKLKSLSITWTNVSGPIPDFLAQLKNLDSLYLSFNNLSGHIPASLSRL 509
Query: 95 KSQLLTYL-------TTPTSMVFFQTTHLVNLFIA 122
+ L L P S FF+ NLF++
Sbjct: 510 PNLLSLNLDRNKLTGPIPASFGFFKKPG-PNLFLS 611
>TC20069 similar to UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3,
partial (14%)
Length = 517
Score = 45.1 bits (105), Expect = 5e-06
Identities = 25/83 (30%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Frame = +3
Query: 15 WSNALL-PLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNL 73
WS P R + L+++ L+G L + + L L+ + L +N S P+P + + L
Sbjct: 219 WSGVTCDPRRHVIALNLTGLSLTGTLSADVAHLPFLSNLSLADNGLSGPIPPSLSAVTGL 398
Query: 74 TTLNLRKCGLIGTFPQNIFQIKS 96
LNL G GTFP + +K+
Sbjct: 399 RFLNLSNNGFNGTFPSELSVLKN 467
Score = 43.9 bits (102), Expect = 1e-05
Identities = 29/86 (33%), Positives = 41/86 (46%)
Frame = +3
Query: 3 LDGISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSP 62
L G+S+T S + L L LS++D LSGP+ SL+ + L + L N F+
Sbjct: 267 LTGLSLTGT---LSADVAHLPFLSNLSLADNGLSGPIPPSLSAVTGLRFLNLSNNGFNGT 437
Query: 63 VPQTFANFKNLTTLNLRKCGLIGTFP 88
P + KNL L+L L G P
Sbjct: 438 FPSELSVLKNLEVLDLYNNNLTGVLP 515
>TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhibiting
protein, partial (93%)
Length = 1195
Score = 44.3 bits (103), Expect = 8e-06
Identities = 24/78 (30%), Positives = 40/78 (50%)
Frame = +3
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
A+ L +L+ L ++ ++SGP+ L+ L NL + L NN + P+P + + NL L
Sbjct: 339 AIAKLTNLRWLILTWTNISGPIPDFLSELPNLQWLHLSFNNLTGPIPSSLSKLPNLIDLR 518
Query: 78 LRKCGLIGTFPQNIFQIK 95
L + L G P + K
Sbjct: 519 LDRNRLTGPIPDSFGSFK 572
Score = 39.3 bits (90), Expect = 3e-04
Identities = 18/60 (30%), Positives = 32/60 (53%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L+GP+ ++ +L NL ++L N S P+P + NL L+L L G P ++ ++
Sbjct: 318 LTGPIQPAIAKLTNLRWLILTWTNISGPIPDFLSELPNLQWLHLSFNNLTGPIPSSLSKL 497
>TC11841 weakly similar to GB|BAC57345.1|28411900|AP005473 receptor protein
kinase CLAVATA1 precursor-like porotein {Oryza sativa
(japonica cultivar-group);} , partial (11%)
Length = 504
Score = 43.9 bits (102), Expect = 1e-05
Identities = 26/75 (34%), Positives = 38/75 (50%)
Frame = +2
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L KLSMS+ +SG L + ++L + + N FSSP+P NF +L L+L
Sbjct: 95 LTKLVKLSMSNNFMSGKLPDNAADFKSLEFLDISNNLFSSPLPPEIGNFGSLQNLSLAGN 274
Query: 82 GLIGTFPQNIFQIKS 96
G P +I + S
Sbjct: 275 NFSGRIPNSISDMAS 319
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 42.7 bits (99), Expect = 2e-05
Identities = 27/68 (39%), Positives = 35/68 (50%)
Frame = +2
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L L + LSG L L L++L + L N S VP +FA KNLT LNL +
Sbjct: 134 LQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRN 313
Query: 82 GLIGTFPQ 89
L G P+
Sbjct: 314 RLHGAIPE 337
Score = 39.3 bits (90), Expect = 3e-04
Identities = 20/70 (28%), Positives = 36/70 (50%)
Frame = +2
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L+ + +S+ LSG + +S L+NLT++ L N +P+ L L L +
Sbjct: 206 LKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWEN 385
Query: 82 GLIGTFPQNI 91
G+ PQ++
Sbjct: 386 NFTGSIPQSL 415
Score = 35.0 bits (79), Expect = 0.005
Identities = 18/55 (32%), Positives = 29/55 (52%)
Frame = +2
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQ 89
L GP+ SL + E+LT I +G+N + +P+ LT + + L G FP+
Sbjct: 533 LFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPE 697
Score = 32.0 bits (71), Expect = 0.040
Identities = 19/60 (31%), Positives = 27/60 (44%)
Frame = +2
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
L L L ++ D LSG + + N+ I L N S P+P T NF ++ L L
Sbjct: 629 LFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLL 808
Score = 26.2 bits (56), Expect = 2.2
Identities = 10/39 (25%), Positives = 17/39 (42%)
Frame = +2
Query: 57 NNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIK 95
NN+ +P N L + CGL G P + +++
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQ 139
>BP053628
Length = 567
Score = 41.6 bits (96), Expect = 5e-05
Identities = 27/95 (28%), Positives = 45/95 (46%), Gaps = 5/95 (5%)
Frame = +1
Query: 15 WSNALLP-----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFAN 69
W+ LP L L+ L +S L G + + + NL + LG+N + +P F +
Sbjct: 22 WNWGSLPSSIHRLYALEALDLSSNYLYGSIPPKIFTIGNLQTLRLGDNFLNGTIPTLFNS 201
Query: 70 FKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYLTT 104
NL L+L+ L G FP +I I + + ++T
Sbjct: 202 SSNLKVLSLKNNRLTGQFPSSILSITTLIEIDMST 306
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 40.8 bits (94), Expect = 9e-05
Identities = 20/82 (24%), Positives = 41/82 (49%)
Frame = +1
Query: 15 WSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLT 74
W + + ++ + + +LSG L L L +L + L ENN +P+ N ++L
Sbjct: 298 WFHVTCQDNSVTRVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLI 477
Query: 75 TLNLRKCGLIGTFPQNIFQIKS 96
+L+L + G+ P ++ +K+
Sbjct: 478 SLDLYHNNVSGSIPSSLGNLKN 543
>TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (70%)
Length = 859
Score = 39.7 bits (91), Expect = 2e-04
Identities = 22/80 (27%), Positives = 36/80 (44%)
Frame = +1
Query: 23 RDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG 82
RDL +L + + L+GP+ + RL+ L ++ L N +P K+LT L L
Sbjct: 7 RDLTRLDLHNNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNS 186
Query: 83 LIGTFPQNIFQIKSQLLTYL 102
G P+ + + YL
Sbjct: 187 FKGEIPKELANLPDLRYLYL 246
Score = 32.7 bits (73), Expect = 0.023
Identities = 19/56 (33%), Positives = 26/56 (45%)
Frame = +1
Query: 48 NLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYLT 103
+LT + L N + P+P K L LNLR L P I ++KS YL+
Sbjct: 10 DLTRLDLHNNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLS 177
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 38.5 bits (88), Expect = 4e-04
Identities = 20/64 (31%), Positives = 32/64 (49%)
Frame = +2
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L + +S +L+G + + L +L+++ L N S PVP +LTTL+L
Sbjct: 1706 LTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFT 1885
Query: 85 GTFP 88
GT P
Sbjct: 1886 GTVP 1897
Score = 36.6 bits (83), Expect = 0.002
Identities = 18/69 (26%), Positives = 34/69 (49%)
Frame = +2
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L+G + L + L ++ +N F P+P+ ++LT + + L G P +FQ+
Sbjct: 1235 LTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQL 1414
Query: 95 KSQLLTYLT 103
S +T L+
Sbjct: 1415 PSVTITELS 1441
Score = 35.0 bits (79), Expect = 0.005
Identities = 22/71 (30%), Positives = 34/71 (46%), Gaps = 11/71 (15%)
Frame = +2
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVP-----------QTFANF 70
L DL L++S ++SGP+ + + +LT + L NNF+ VP +TFA
Sbjct: 1769 LMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGN 1948
Query: 71 KNLTTLNLRKC 81
NL + C
Sbjct: 1949 PNLCFPHRASC 1981
Score = 33.1 bits (74), Expect = 0.018
Identities = 23/92 (25%), Positives = 44/92 (47%), Gaps = 1/92 (1%)
Frame = +2
Query: 6 ISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLT-RLENLTVIVLGENNFSSPVP 64
IS+ + + + L L L+ L++S SG ++T + L + +N+FS P+P
Sbjct: 422 ISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLP 601
Query: 65 QTFANFKNLTTLNLRKCGLIGTFPQNIFQIKS 96
+ + L L+L GT P++ + +S
Sbjct: 602 EEIVKLEKLKYLHLAGNYFSGTIPESYSEFQS 697
Score = 26.6 bits (57), Expect = 1.7
Identities = 17/67 (25%), Positives = 26/67 (38%)
Frame = +2
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L+ ++D GP+ + +LT I + N PVP ++T L L
Sbjct: 1277 LKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLN 1456
Query: 85 GTFPQNI 91
G P I
Sbjct: 1457 GELPSVI 1477
>TC14163
Length = 1712
Score = 38.5 bits (88), Expect = 4e-04
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Frame = +1
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L+ L + LSG + L + L + L N FS VP +F N + LNL L+
Sbjct: 856 LRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLV 1035
Query: 85 GTFPQ-NIFQIKS 96
FP+ N+ I+S
Sbjct: 1036DPFPEMNVKGIES 1074
Score = 28.1 bits (61), Expect = 0.58
Identities = 17/57 (29%), Positives = 26/57 (44%), Gaps = 1/57 (1%)
Frame = +1
Query: 36 SGPLDSSLTRLENLT-VIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNI 91
SG + SL++++NL +L N S P P L + + L G P+NI
Sbjct: 379 SGTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENI 549
>TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Arabidopsis
thaliana;}, partial (77%)
Length = 1318
Score = 38.1 bits (87), Expect = 6e-04
Identities = 25/84 (29%), Positives = 35/84 (40%), Gaps = 8/84 (9%)
Frame = +1
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
+ KLS+++ L G + L NL + L N + P+P + NL LNL L
Sbjct: 559 IYKLSLNNLALHGTISPFLANCTNLQALDLSSNFLTGPIPPDLQSLVNLAVLNLSANRLE 738
Query: 85 GTFPQ--------NIFQIKSQLLT 100
G P NI + LLT
Sbjct: 739 GEIPPQLTMCAYLNIIDLHQNLLT 810
Score = 25.8 bits (55), Expect = 2.9
Identities = 10/35 (28%), Positives = 17/35 (48%)
Frame = +1
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFAN 69
L+GP+ L L L+ + N + P+P + N
Sbjct: 805 LTGPIPQQLGLLVRLSAFDVSNNRLAGPIPSSLTN 909
>TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor family 3,
partial (24%)
Length = 565
Score = 38.1 bits (87), Expect = 6e-04
Identities = 23/69 (33%), Positives = 34/69 (48%)
Frame = +2
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
+L L L +S+++ LSG + + L L + L NN S +P + N LTTL
Sbjct: 236 SLSTLTGLTDMSLNENHLSGEIPDAFQSLTQLINLDLSTNNLSGELPPSVENLSALTTLR 415
Query: 78 LRKCGLIGT 86
L+ L GT
Sbjct: 416 LQNNQLSGT 442
>AU240163
Length = 300
Score = 37.4 bits (85), Expect = 0.001
Identities = 20/64 (31%), Positives = 34/64 (52%)
Frame = +1
Query: 31 SDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQN 90
+D +SG + +SLT L +L + L N S P+P+ F + + L+ L + G P +
Sbjct: 61 ADNGISGAIPASLTGLRSLMHLDLRNNRLSGPIPRDFGSLRMLSRALLSGNSISGPIPSS 240
Query: 91 IFQI 94
I +I
Sbjct: 241 ISRI 252
Score = 35.4 bits (80), Expect = 0.004
Identities = 19/63 (30%), Positives = 31/63 (49%)
Frame = +1
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
+L LR L L + + LSGP+ L L+ +L N+ S P+P + + L L+
Sbjct: 94 SLTGLRSLMHLDLRNNRLSGPIPRDFGSLRMLSRALLSGNSISGPIPSSISRIYRLADLD 273
Query: 78 LRK 80
L +
Sbjct: 274 LSR 282
>TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-like protein,
partial (19%)
Length = 570
Score = 36.6 bits (83), Expect = 0.002
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 1/88 (1%)
Frame = +1
Query: 14 EWSNALLPLRDLQKLSMSDCDLSGPLDS-SLTRLENLTVIVLGENNFSSPVPQTFANFKN 72
+W + + + D L+G + +LTRL+ L V+ L N+ + P+P + N
Sbjct: 217 QWQGVKCSQGRVFRFVLQDLSLAGTFPADTLTRLDQLRVLTLRNNSLTGPIPD-LSPLTN 393
Query: 73 LTTLNLRKCGLIGTFPQNIFQIKSQLLT 100
L +L L + G FP +I + +LLT
Sbjct: 394 LKSLFLDRNHFSGAFPPSILSL-HRLLT 474
Score = 28.1 bits (61), Expect = 0.58
Identities = 17/42 (40%), Positives = 23/42 (54%)
Frame = +1
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNF 59
++L L L LS+S +LSG L L L LT + L N+F
Sbjct: 445 SILSLHRLLTLSLSHNNLSGQLPVQLNLLXRLTSLRLDSNSF 570
>BP033575
Length = 515
Score = 36.6 bits (83), Expect = 0.002
Identities = 26/76 (34%), Positives = 35/76 (45%)
Frame = +3
Query: 3 LDGISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSP 62
LDG + G + LL L+ L LS++ +G L SL L +L + L NNF P
Sbjct: 282 LDGFGLG--GELKFHTLLDLKMLSNLSLAGNHFTGRLPPSLGTLTSLQHLDLSRNNFYGP 455
Query: 63 VPQTFANFKNLTTLNL 78
+P L LNL
Sbjct: 456 IPARINELWGLNYLNL 503
>AV415057
Length = 406
Score = 35.4 bits (80), Expect = 0.004
Identities = 20/57 (35%), Positives = 29/57 (50%)
Frame = +3
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNI 91
LSG + + +L L + L NNFS +P +N NL TL+L L G P ++
Sbjct: 189 LSGSIPIEIGQLSVLHQLDLKNNNFSGNIPVQISNLTNLETLDLSGNHLSGEIPDSL 359
>TC9071 similar to UP|Q9LT85 (Q9LT85) Similarity to receptor protein
kinase, partial (15%)
Length = 872
Score = 33.9 bits (76), Expect = 0.011
Identities = 21/68 (30%), Positives = 30/68 (43%)
Frame = +1
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L G LDS + LT + NNFS VP F L++ G FP ++ +
Sbjct: 526 LYGILDS----IPELTFFHVNSNNFSGAVPSEILKFTYFYELDISNNKFSGEFPMDVLKF 693
Query: 95 KSQLLTYL 102
S+ L +L
Sbjct: 694 SSKQLIFL 717
>AV408059
Length = 280
Score = 33.9 bits (76), Expect = 0.011
Identities = 16/46 (34%), Positives = 23/46 (49%)
Frame = +1
Query: 49 LTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
L V+ + NNFS +P N K L L+L GTFP ++ +
Sbjct: 52 LVVLNMTRNNFSGEIPMKIGNMKCLQNLDLSWNNFSGTFPSSLVNL 189
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,352,862
Number of Sequences: 28460
Number of extensions: 28196
Number of successful extensions: 228
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 226
length of query: 139
length of database: 4,897,600
effective HSP length: 81
effective length of query: 58
effective length of database: 2,592,340
effective search space: 150355720
effective search space used: 150355720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 50 (23.9 bits)
Medicago: description of AC144474.15


