

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144389.13 - phase: 0
(596 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
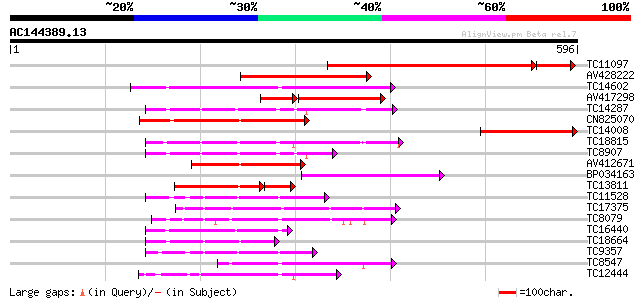
Score E
Sequences producing significant alignments: (bits) Value
TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-depende... 422 e-133
AV428222 248 3e-66
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 179 9e-46
AV417298 125 5e-42
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 162 2e-40
CN825070 152 2e-37
TC14008 similar to UP|Q8RY07 (Q8RY07) Serine/threonine protein k... 146 8e-36
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 140 6e-34
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 134 4e-32
AV412671 132 2e-31
BP034163 129 2e-30
TC13811 homologue to UP|O24431 (O24431) Calmodulin-like domain p... 84 8e-25
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 104 4e-23
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 103 1e-22
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 100 5e-22
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 99 2e-21
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 97 7e-21
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 96 2e-20
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 92 3e-19
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 84 5e-17
>TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-dependent protein
kinase homolog|CaM kinase homolog|MCK1, partial (36%)
Length = 949
Score = 422 bits (1086), Expect(2) = e-133
Identities = 208/219 (94%), Positives = 213/219 (96%)
Frame = +2
Query: 335 GVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKR 394
GVITYILLCGSRPFWARTESGIFR VLRADPNFDDLPWPSV+PE KDFVKRLLNKDYRKR
Sbjct: 20 GVITYILLCGSRPFWARTESGIFRAVLRADPNFDDLPWPSVTPEAKDFVKRLLNKDYRKR 199
Query: 395 MTAAQALTHPWLRDESRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRA 454
MTAAQALTHPWLRD+SRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALT++QLVYLRA
Sbjct: 200 MTAAQALTHPWLRDDSRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTEEQLVYLRA 379
Query: 455 QFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAAT 514
QFRLLEPNRDGH+SLDNFKMALARHATDAMRESRVLDIIH MEPLAYRKMDFEEFCAAAT
Sbjct: 380 QFRLLEPNRDGHISLDNFKMALARHATDAMRESRVLDIIHMMEPLAYRKMDFEEFCAAAT 559
Query: 515 STYQLEALDGWEDIACAAFEHFESEGNRVISIEELAREL 553
STYQLEALD WEDIA AAFEHFE EGNRVISIEELAREL
Sbjct: 560 STYQLEALDRWEDIASAAFEHFEREGNRVISIEELAREL 676
Score = 70.1 bits (170), Expect(2) = e-133
Identities = 32/41 (78%), Positives = 36/41 (87%)
Frame = +3
Query: 554 NLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSSLPR 594
NLGPSAYS+LRDWIR DGKL+LLGY KFL GVTLRS +P+
Sbjct: 678 NLGPSAYSILRDWIRRRDGKLNLLGYPKFLPGVTLRSPVPQ 800
>AV428222
Length = 416
Score = 248 bits (632), Expect = 3e-66
Identities = 115/138 (83%), Positives = 129/138 (93%)
Frame = +3
Query: 243 RYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRP 302
RY E+DAKAI+LQIL+V AFCHL GVVHRDLKPENFLF S+ D+ MK+IDFGLSDF+RP
Sbjct: 3 RYPEDDAKAILLQILNVVAFCHLHGVVHRDLKPENFLFVSKEADSVMKVIDFGLSDFVRP 182
Query: 303 EERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLR 362
++RLNDIVGSAYYVAPEVLHRSYSVEAD+WSIGVI+YILLCGSRPFWARTESGIFR+VLR
Sbjct: 183 DQRLNDIVGSAYYVAPEVLHRSYSVEADLWSIGVISYILLCGSRPFWARTESGIFRSVLR 362
Query: 363 ADPNFDDLPWPSVSPEGK 380
A+PNFDD PWPS+SPE K
Sbjct: 363 ANPNFDDSPWPSISPEAK 416
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 179 bits (455), Expect = 9e-46
Identities = 101/280 (36%), Positives = 159/280 (56%), Gaps = 2/280 (0%)
Frame = +2
Query: 128 QSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAI 187
QS ++ + ++Y+L +E+GRG FG + AVK+I KS + +
Sbjct: 20 QSRERK*AMCEALKSQYQLCEEIGRGRFGTIFRCFHP---ISTDTFAVKLIDKSLLADST 190
Query: 188 AIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEE 247
+ E K + LS H ++++ D ED + + +V+ELC+ LLDRI++ G E
Sbjct: 191 DRHCLENEPKYMSLLSPHPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPE 370
Query: 248 -DAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERL 306
+A ++ Q+L A CH GV HRD+KP+N LF D+KL DFG +++ R+
Sbjct: 371 VEAAGLMKQLLEAVAHCHRLGVAHRDVKPDNVLFGG---GGDLKLADFGSAEWFGDGRRM 541
Query: 307 NDIVGSAYYVAPEVLH-RSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADP 365
+ +VG+ YYVAPEVL R Y + D+WS GVI YI+L G+ PF+ + + IF V+R +
Sbjct: 542 SGVVGTPYYVAPEVLMGREYGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNL 721
Query: 366 NFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPW 405
F + +VSP KD +++++ +D R++A QAL HPW
Sbjct: 722 RFPSRIFRNVSPAAKDLLRKMICRDPSNRISAEQALRHPW 841
>AV417298
Length = 424
Score = 125 bits (315), Expect(2) = 5e-42
Identities = 59/92 (64%), Positives = 70/92 (75%)
Frame = +1
Query: 304 ERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRA 363
++ DIVGSAYYVAPEVL R E+D+WSIGVITYILLCG RPFW +TE GIF+ VLR
Sbjct: 148 KKFQDIVGSAYYVAPEVLKRKSGPESDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRN 327
Query: 364 DPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRM 395
P+F PWP++S KDFVK+LL KD R R+
Sbjct: 328 KPDFRRKPWPTISNAAKDFVKKLLVKDPRARL 423
Score = 62.8 bits (151), Expect(2) = 5e-42
Identities = 27/39 (69%), Positives = 32/39 (81%)
Frame = +2
Query: 264 HLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRP 302
HL G+VHRD+KPENFLF S ED+ +K DFGLSDFI+P
Sbjct: 2 HLHGLVHRDMKPENFLFKSNREDSALKATDFGLSDFIKP 118
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 162 bits (409), Expect = 2e-40
Identities = 96/272 (35%), Positives = 151/272 (55%), Gaps = 7/272 (2%)
Frame = +3
Query: 143 KYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKAL 202
KYE+G+ +G+G+F R ++ VA+K+I K K+ ++ ++REV +++ L
Sbjct: 429 KYEMGRVLGQGNFAKVYHGRNLA---TNENVAIKVIKKEKLKKDRLVKQIKREVSVMR-L 596
Query: 203 SGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAF 262
H H+++ + +++VME +GGEL ++ G+ E+DA+ Q++S F
Sbjct: 597 VRHPHIVELKEVMATKGKIFLVMEYVKGGELFTKV--NKGKLNEDDARKYFQQLISAVDF 770
Query: 263 CHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIV-----GSAYYVA 317
CH +GV HRDLKPEN L E+ D+K+ DFGLS PE+R +D + G+ YVA
Sbjct: 771 CHSRGVTHRDLKPENLLL---DENEDLKVSDFGLSAL--PEQRRDDGMLVTPCGTPAYVA 935
Query: 318 PEVLHRS--YSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSV 375
PEVL + +ADIWS GVI Y LL G PF I+R +A+ F + W +
Sbjct: 936 PEVLKKKGYDGSKADIWSCGVILYALLSGYLPFQGENVMRIYRKAFKAEYEFPE--W--I 1103
Query: 376 SPEGKDFVKRLLNKDYRKRMTAAQALTHPWLR 407
SP+ K+ + LL D KR + + ++ PW +
Sbjct: 1104SPQAKNLISNLLVADPEKRYSIPEIISDPWFQ 1199
>CN825070
Length = 634
Score = 152 bits (383), Expect = 2e-37
Identities = 79/179 (44%), Positives = 110/179 (61%)
Frame = +3
Query: 137 SKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREV 196
++N Y G+++G+G FG T R A K I K K+ +DV RE+
Sbjct: 108 TENIREVYTFGRKLGQGQFGTTYLCRHNSTGCT---FACKSIPKRKLLCKEDYDDVWREI 278
Query: 197 KLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQI 256
+++ LS H H+++ H EDA +V+IVME+CEGGEL DRI+ +G +Y+E +A ++ I
Sbjct: 279 QIMHHLSEHPHVVRIHGTYEDAASVHIVMEICEGGELFDRIVQKG-QYSEREAAKLIRTI 455
Query: 257 LSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYY 315
+ V CH GV+HRDLKPENFLF + EDA +K DFGLS F +P E D+VGS YY
Sbjct: 456 VEVVEACHSLGVMHRDLKPENFLFDTVEEDAKLKTTDFGLSVFYKPGETFGDVVGSPYY 632
>TC14008 similar to UP|Q8RY07 (Q8RY07) Serine/threonine protein kinase pk23,
partial (17%)
Length = 619
Score = 146 bits (369), Expect = 8e-36
Identities = 69/101 (68%), Positives = 82/101 (80%)
Frame = +2
Query: 496 MEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELARELNL 555
ME L+Y+K+DFEEFCAAA S YQLEA W+ IA AF +F+ GNRVIS+EELA+E+NL
Sbjct: 2 MESLSYKKLDFEEFCAAAISVYQLEAHPEWDKIAATAFAYFDETGNRVISVEELAQEMNL 181
Query: 556 GPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSSLPRPR 596
GPS YS++ DWIR +DGKLSL GYTKFLHGVT+RSS R R
Sbjct: 182 GPSTYSLMNDWIRKSDGKLSLFGYTKFLHGVTIRSSNSRHR 304
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 140 bits (353), Expect = 6e-34
Identities = 89/280 (31%), Positives = 147/280 (51%), Gaps = 8/280 (2%)
Frame = +2
Query: 143 KYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKAL 202
KYE+G+ +G+G A K + VA+K++SK+++ ++ ++RE+ +++ L
Sbjct: 35 KYEMGRVLGKGTLAKVYFA---KEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMR-L 202
Query: 203 SGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAF 262
H +++ + ++ +ME GGEL ++ G+ ++ A+ Q++S +
Sbjct: 203 VRHPNIVNLKEVMATKTKIFFIMEYIRGGELFAKVAK--GKLKDDLARRYFQQLISAVDY 376
Query: 263 CHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLS---DFIRPEERLNDIVGSAYYVAPE 319
CH +GV HRDLKPEN L E+ ++K+ DFGLS + +R + L+ G+ YVAPE
Sbjct: 377 CHSRGVSHRDLKPENLLL---DENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPE 547
Query: 320 VLHRS--YSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSP 377
VL + + D WS GVI Y LL G PF ++ VLRA+ F PW SP
Sbjct: 548 VLRKKGYDGFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLRAEFQFP--PW--FSP 715
Query: 378 EGKDFVKRLLNKDYRKRMTAAQALTHPWLR---DESRPIP 414
E K + ++L D +R+T + + W + S PIP
Sbjct: 716 ESKKLISKILVADPNRRITISSIMRVSWFQKGFSASIPIP 835
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 134 bits (337), Expect = 4e-32
Identities = 78/209 (37%), Positives = 122/209 (58%), Gaps = 7/209 (3%)
Frame = +3
Query: 143 KYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKAL 202
KYE+GK +G+G+F R + ++ VA+K+I K ++ ++ ++REV +++ L
Sbjct: 339 KYEIGKILGQGNFAKVYHGRNME---TNESVAIKVIKKERLKKERLVKQIKREVSVMR-L 506
Query: 203 SGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAF 262
H H+++ + +++V+E +GGEL ++ G+ TE A+ Q++S F
Sbjct: 507 VRHPHIVELKEVMATKTKIFMVVEYVKGGELFAKLTK--GKMTEVAARKYFQQLISAVDF 680
Query: 263 CHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIV-----GSAYYVA 317
CH +GV HRDLKPEN L ++ D+K+ DFGLS PE+R +D + G+ YVA
Sbjct: 681 CHSRGVTHRDLKPENLLL---DDNEDLKVSDFGLSSL--PEQRRSDGMLLTPCGTPAYVA 845
Query: 318 PEVLHRS--YSVEADIWSIGVITYILLCG 344
PEVL + +ADIWS GVI Y LLCG
Sbjct: 846 PEVLKKKGYDGSKADIWSCGVILYALLCG 932
>AV412671
Length = 359
Score = 132 bits (331), Expect = 2e-31
Identities = 65/120 (54%), Positives = 84/120 (69%)
Frame = +3
Query: 192 VRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKA 251
VRREV++++ L H +++ D ED N V++VMELCEGGEL DRI++RG YTE A A
Sbjct: 3 VRREVEIMRHLPRHPNIVWLKDTYEDDNAVHLVMELCEGGELFDRIVARG-HYTERAAAA 179
Query: 252 IVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVG 311
+ I+ V CH GV+HRDLKPENFLF ++ E A +K IDFGLS F +P E N+IVG
Sbjct: 180 VTKTIVEVVQMCHKHGVMHRDLKPENFLFANKKETAALKAIDFGLSVFFKPGETFNEIVG 359
>BP034163
Length = 477
Score = 129 bits (323), Expect = 2e-30
Identities = 64/153 (41%), Positives = 93/153 (59%), Gaps = 2/153 (1%)
Frame = +1
Query: 307 NDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPN 366
N+I GS YY+APEVL R Y E DIWS GVI YILLCG PFWA TE + + ++R+ +
Sbjct: 7 NEIXGSPYYMAPEVLRRHYGPEVDIWSAGVILYILLCGVPPFWAETEQVVAQAIIRSVVD 186
Query: 367 FDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWL--RDESRPIPLDILIYKLVK 424
F PWP VS KD VK++LN D ++R+TA + L HPWL ++ + L + +K
Sbjct: 187 FKRDPWPKVSDNAKDLVKKMLNPDPKRRLTAQEVLDHPWLVHAKKAPNVSLGETVKARLK 366
Query: 425 SYLHATPFKRAAVKALSKALTDDQLVYLRAQFR 457
+ K+ A++ +++ LT ++ LR F+
Sbjct: 367 QFSVMNKLKKRALRVIAEHLTVEEAAGLREGFQ 465
>TC13811 homologue to UP|O24431 (O24431) Calmodulin-like domain protein
kinase isoenzyme gamma , partial (27%)
Length = 573
Score = 84.0 bits (206), Expect(2) = 8e-25
Identities = 41/94 (43%), Positives = 63/94 (66%)
Frame = +1
Query: 174 AVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGEL 233
A K ISK K+ S ED++RE+++++ L G ++++F A ED +V++VMELC GGEL
Sbjct: 52 ACKSISKRKLVSKSDKEDIKREIQIMQHLIGQPNIVEFKGAYEDKQSVHVVMELCAGGEL 231
Query: 234 LDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQG 267
DRI+++ G Y+E A +I QI++V CH G
Sbjct: 232 FDRIIAK-GHYSERAAASICRQIVNVVHVCHFMG 330
Score = 47.0 bits (110), Expect(2) = 8e-25
Identities = 22/33 (66%), Positives = 26/33 (78%)
Frame = +2
Query: 268 VVHRDLKPENFLFTSRSEDADMKLIDFGLSDFI 300
V+HRDLKPENFL +S+ E A +K DFGLS FI
Sbjct: 332 VMHRDLKPENFLLSSKDEKALLKATDFGLSVFI 430
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 104 bits (260), Expect = 4e-23
Identities = 73/197 (37%), Positives = 108/197 (54%), Gaps = 3/197 (1%)
Frame = +3
Query: 143 KYELGKEVGRGHFGHTCSARAKK-GDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKA 201
+YE KE+G G+FG AR K G+L VAVK I + K E+V+RE+ ++
Sbjct: 75 RYEPLKELGSGNFGVARLARDKNTGEL----VAVKYIERGKKID----ENVQREIINHRS 230
Query: 202 LSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAA 261
L H ++I+F + ++ IV+E GGEL +RI S GR++E++A+ Q++S +
Sbjct: 231 LR-HPNIIRFKEVLLTPTHLAIVLEYASGGELFERICS-AGRFSEDEARYFFQQLISGVS 404
Query: 262 FCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVL 321
+CH + HRDLK EN L + +K+ DFG S + VG+ Y+APEVL
Sbjct: 405 YCHSMEICHRDLKLENTLLDG-NPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVL 581
Query: 322 HRSY--SVEADIWSIGV 336
R AD+WS GV
Sbjct: 582 SRKEYDGKVADVWSCGV 632
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 103 bits (256), Expect = 1e-22
Identities = 70/238 (29%), Positives = 123/238 (51%), Gaps = 2/238 (0%)
Frame = +3
Query: 175 VKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELL 234
VK+ S K TS ++ + +E+ LL S H ++++++ + ++ + +E GG +
Sbjct: 24 VKVFSDDK-TSKECLKQLNQEINLLNQFS-HPNIVQYYGSELGEESLSVYLEYVSGGSI- 194
Query: 235 DRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDF 294
++L G + E + QI+S A+ H + VHRD+K N L E +KL DF
Sbjct: 195 HKLLQEYGAFKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGE---IKLADF 365
Query: 295 GLSDFIRPEERLNDIVGSAYYVAPEVLHRS--YSVEADIWSIGVITYILLCGSRPFWART 352
G+S I + GS Y++APEV+ + Y + DI S+G T + + S+P W++
Sbjct: 366 GMSKHINSAASMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGC-TILEMATSKPPWSQF 542
Query: 353 ESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDES 410
E + + ++P +S + K+F+K+ L +D R TA L HP++RD+S
Sbjct: 543 EGVAAIFKIGNSKDMPEIP-EHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIRDQS 713
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 100 bits (250), Expect = 5e-22
Identities = 85/292 (29%), Positives = 138/292 (47%), Gaps = 35/292 (11%)
Frame = +3
Query: 150 VGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLI 209
+GRG +G CS + ++ VAVK I+ + + + + RE+KLL+ L H+++I
Sbjct: 309 IGRGAYGIVCSLLNTE---TNELVAVKKIANA-FDNHMDAKRTLREIKLLRHLD-HENVI 473
Query: 210 KFHDAC-----EDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAFCH 264
D + +VYI EL + L +I+ +EE + + Q+L + H
Sbjct: 474 ALRDVIPPPLRREFTDVYITTELMDTD--LHQIIRSNQGLSEEHCQYFLYQVLRGLKYIH 647
Query: 265 LQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRS 324
++HRDLKP N L S + D+K+IDFGL+ + + + V + +Y APE+L S
Sbjct: 648 SANIIHRDLKPSNLLLNS---NCDLKIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNS 818
Query: 325 --YSVEADIWSIGVITYILLCGSRPFW-----------------ARTESGI-------FR 358
Y+ D+WS+G I ++ L +P + TE+ + R
Sbjct: 819 SDYTSAIDVWSVGCI-FMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVR 995
Query: 359 TVLRADPNFDDLP----WPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWL 406
+R P + P +P V P D V ++L D KR+T QAL HP+L
Sbjct: 996 RYIRQLPQYPRQPLTKVFPHVHPMAMDLVDKMLTIDPTKRITVEQALAHPYL 1151
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 99.0 bits (245), Expect = 2e-21
Identities = 58/156 (37%), Positives = 87/156 (55%), Gaps = 1/156 (0%)
Frame = +1
Query: 143 KYELGKEVGRGHFGHTCSARAKKGDLKD-QPVAVKIISKSKMTSAIAIEDVRREVKLLKA 201
KY+L + +GRG+F A + L D VAVK+I KSK A + RE+ ++
Sbjct: 88 KYQLTRFLGRGNFAKVYQAVS----LTDGTTVAVKMIDKSKTVDASMEPRIVREIDAMRR 255
Query: 202 LSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAA 261
L H +++K H+ +Y++++ GGEL +I SR GR E A+ Q++S
Sbjct: 256 LQHHPNILKIHEVMATRTKIYLIVDYAGGGELFSKI-SRRGRLPEPLARRYFQQLVSALC 432
Query: 262 FCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLS 297
FCH GV HRDLKP+N L + + ++K+ DFGLS
Sbjct: 433 FCHRNGVAHRDLKPQNLLLDA---EGNLKVSDFGLS 531
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 97.1 bits (240), Expect = 7e-21
Identities = 53/141 (37%), Positives = 81/141 (56%)
Frame = +3
Query: 143 KYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKAL 202
KYE+G+ +G G F AR + Q VAVK+I+K K+ V+REV ++ L
Sbjct: 90 KYEVGRLLGCGAFAKVYHARNIE---TGQSVAVKVINKKKVIGTGLTGHVKREVSIMSRL 260
Query: 203 SGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAF 262
H ++++ H+ +Y VME +GGEL RI ++G R++E+ A+ Q++S +
Sbjct: 261 R-HPNIVRLHEVLATKTKIYFVMEFAKGGELFARISTKG-RFSEDLARRYFQQLISAVGY 434
Query: 263 CHLQGVVHRDLKPENFLFTSR 283
CH +GV HRDLKPEN L +
Sbjct: 435 CHSRGVFHRDLKPENLLLDDK 497
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 95.9 bits (237), Expect = 2e-20
Identities = 62/181 (34%), Positives = 99/181 (54%)
Frame = +1
Query: 143 KYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKAL 202
+Y+L +++G G+FG AR + + VAVK I + E+V+RE+ ++L
Sbjct: 358 RYDLVRDIGSGNFG---VARLMQDKQTKELVAVKYIERGDKID----ENVKREIINHRSL 516
Query: 203 SGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAF 262
H ++++F + ++ IVME GGEL +RI + GR+TE++A+ Q++S ++
Sbjct: 517 R-HPNIVRFKEVILTPTHLAIVMEYASGGELFERICN-AGRFTEDEARFFFQQLISGVSY 690
Query: 263 CHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLH 322
CH V HRDLK EN L S +K+ DFG S + VG+ Y+APEVL
Sbjct: 691 CHAMQVCHRDLKLENTLLDG-SPTPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLS 867
Query: 323 R 323
+
Sbjct: 868 K 870
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 91.7 bits (226), Expect = 3e-19
Identities = 67/216 (31%), Positives = 101/216 (46%), Gaps = 28/216 (12%)
Frame = +1
Query: 219 NNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENF 278
N+VYI EL + L +I+ +EE + + QIL + H V+HRDLKP N
Sbjct: 58 NDVYIAYELMDTD--LHQIIRSNQGLSEEHCQYFLYQILRGLKYIHSANVLHRDLKPSNL 231
Query: 279 LFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRS--YSVEADIWSIGV 336
L + + D+K+ DFGL+ + + + V + +Y APE+L S Y+ D+WS+G
Sbjct: 232 LLNA---NCDLKICDFGLARVTSETDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGC 402
Query: 337 ITYILLCGSRPFWARTESGIFRTVLR--ADPNFDDL------------------------ 370
I L+ F R R ++ P+ DDL
Sbjct: 403 IFMELMDRKPLFPGRDHVHQLRLLMELIGTPSEDDLGFLNENAKRYIRQLPPYRRQSFQE 582
Query: 371 PWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWL 406
+P V PE D V+++L D RKR+T +AL HP+L
Sbjct: 583 KFPQVHPEAIDLVEKMLTFDPRKRITVEEALAHPYL 690
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 84.3 bits (207), Expect = 5e-17
Identities = 68/222 (30%), Positives = 109/222 (48%), Gaps = 9/222 (4%)
Frame = +2
Query: 136 YSKNFGAKYELGKEVGRGHFGHTCSARAKKGDL--KDQPVAVKIISKSKMTSAIAIEDVR 193
YS N G Y L +EVG G SA + +D+ VAVK + + + + ED+R
Sbjct: 68 YSLNSG-DYNLLEEVGYG-----ASATVYRAVFLPRDEEVAVKCVDLDRCNANL--EDIR 223
Query: 194 REVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRI-LSRGGRYTEEDAKAI 252
RE + + +L H+++++ H + +++VM G L + ++ + EE +I
Sbjct: 224 REAQTM-SLIDHRNVVRAHWSFVVDRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSI 400
Query: 253 VLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLS----DFIRPEERLND 308
+ + L + H G +HRD+K N L S D +KL DFG+S D + N
Sbjct: 401 LKETLKALEYLHRHGHIHRDVKAGNILLGS---DGQVKLADFGVSASMFDAGERQRNRNT 571
Query: 309 IVGSAYYVAPEVLH--RSYSVEADIWSIGVITYILLCGSRPF 348
VG+ ++APEVL Y+ +AD+WS G+ L G PF
Sbjct: 572 FVGTPCWMAPEVLQPGTGYNFKADVWSFGITALELAHGHAPF 697
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,158,979
Number of Sequences: 28460
Number of extensions: 158284
Number of successful extensions: 2354
Number of sequences better than 10.0: 392
Number of HSP's better than 10.0 without gapping: 1821
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2113
length of query: 596
length of database: 4,897,600
effective HSP length: 95
effective length of query: 501
effective length of database: 2,193,900
effective search space: 1099143900
effective search space used: 1099143900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144389.13


