

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.8 + phase: 0
(396 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
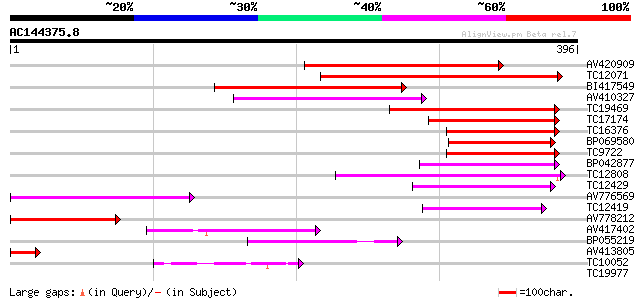
Score E
Sequences producing significant alignments: (bits) Value
AV420909 214 2e-56
TC12071 weakly similar to GB|AAP31968.1|30387605|BT006624 At2g04... 178 1e-45
BI417549 147 3e-36
AV410327 99 2e-21
TC19469 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, par... 97 6e-21
TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120,... 86 1e-17
TC16376 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (17%) 85 2e-17
BP069580 85 2e-17
TC9722 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (16%) 84 3e-17
BP042877 84 4e-17
TC12808 weakly similar to GB|CAA16564.1|2827556|ATT12H17 predict... 84 4e-17
TC12429 similar to UP|Q9FKQ1 (Q9FKQ1) Emb|CAB89401.1 (AT5g65380/... 81 3e-16
AV776569 80 6e-16
TC12419 78 3e-15
AV778212 62 2e-10
AV417402 62 2e-10
BP055219 48 3e-06
AV413805 41 3e-04
TC10052 40 7e-04
TC19977 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (20%) 31 0.31
>AV420909
Length = 421
Score = 214 bits (545), Expect = 2e-56
Identities = 103/139 (74%), Positives = 117/139 (84%)
Frame = +3
Query: 207 SVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFL 266
SVCL+TTTLHYF+ +A+GA+ASTRVSNELGAGNP+ AKGAVRV VI+GI EA+IVS F+
Sbjct: 3 SVCLNTTTLHYFVTYAVGAAASTRVSNELGAGNPKTAKGAVRVVVIVGIVEAIIVSAFFI 182
Query: 267 CFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGA 326
CFRN++G AYSNDKEVVDYV DM P LC SV DS+I ALSG+ARGGGFQ IGAY NLGA
Sbjct: 183 CFRNVLGYAYSNDKEVVDYVADMAPLLCGSVIGDSLIGALSGVARGGGFQQIGAYANLGA 362
Query: 327 YYLVGAPIAYFLGFGLKLN 345
YY+VG PI LGF L LN
Sbjct: 363 YYVVGIPIGLLLGFHLHLN 419
>TC12071 weakly similar to GB|AAP31968.1|30387605|BT006624 At2g04100
{Arabidopsis thaliana;}, partial (32%)
Length = 764
Score = 178 bits (452), Expect = 1e-45
Identities = 86/169 (50%), Positives = 117/169 (68%)
Frame = +3
Query: 218 FIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYS 277
F + +GA+ STRVSNELGAG P+ A+ A+ +I+ +A+I+S+ C R+++G A+S
Sbjct: 15 FYSYGVGAAVSTRVSNELGAGKPKEARDAIFAVIILATLDAIILSSALFCCRHVLGFAFS 194
Query: 278 NDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYF 337
ND EVV V +VP LC+SV DS + LSG+ARG G+Q IGA NL AYY +G P+A
Sbjct: 195 NDLEVVHSVAKIVPLLCLSVCVDSFLGVLSGVARGAGWQKIGAVTNLLAYYAIGIPVALL 374
Query: 338 LGFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERIAE 386
LGF LKLNAKGLW+G LTGS ++LA++T T W+K+A A R +E
Sbjct: 375 LGFVLKLNAKGLWIGILTGSTTQTLVLALLTAFTKWEKQAPLATVRTSE 521
>BI417549
Length = 404
Score = 147 bits (371), Expect = 3e-36
Identities = 69/134 (51%), Positives = 98/134 (72%)
Frame = +1
Query: 144 MKYSPACEKTKIVFSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLET 203
+K S ACEKT+ FS +L + EF + ++PS M C++WW+ E+L ++AGL N +LET
Sbjct: 1 VKCSSACEKTRTPFSKRALSGLGEFFRLSVPSAAMVCVKWWASELLVLLAGLFSNPELET 180
Query: 204 SVLSVCLSTTTLHYFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVST 263
S+LS+CL+ +TLH+ IP+ G +ASTRV+NELGAGNP+AA+ V +A+ + EAVIVST
Sbjct: 181 SILSICLTISTLHFTIPYGFGVAASTRVANELGAGNPKAARVVVSIAMSLAFIEAVIVST 360
Query: 264 LFLCFRNIIGNAYS 277
FR+I+G AYS
Sbjct: 361 ALFGFRHILGYAYS 402
>AV410327
Length = 424
Score = 98.6 bits (244), Expect = 2e-21
Identities = 51/135 (37%), Positives = 80/135 (58%)
Frame = +2
Query: 157 FSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLH 216
FS+ + + EF + + S +M CLE W F+IL ++AGLLP+ +L LS+C + +
Sbjct: 20 FSWQAFSELPEFFKLSAASAVMLCLETWYFQILVLLAGLLPHPELALDSLSICTTISGWV 199
Query: 217 YFIPHAIGASASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAY 276
+ I A+AS RVSNELGA NP++A +V V +I +VI + + L R+II +
Sbjct: 200 FMISVGFNAAASVRVSNELGARNPKSASFSVVVVTLISFIMSVIAALVVLALRDIISYVF 379
Query: 277 SNDKEVVDYVTDMVP 291
+ +EV V+D+ P
Sbjct: 380 TGGEEVAAAVSDLCP 424
>TC19469 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (26%)
Length = 501
Score = 96.7 bits (239), Expect = 6e-21
Identities = 43/119 (36%), Positives = 72/119 (60%)
Frame = +1
Query: 266 LCFRNIIGNAYSNDKEVVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLG 325
L R + ++ D EV + V D+ P L +S+ +S+ LSG++ G G+ ++ AYVN+G
Sbjct: 1 LFLRERLAYVFTLDSEVAEAVGDLSPLLSISILLNSVPPVLSGVSVGAGWPSVVAYVNIG 180
Query: 326 AYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERI 384
YYL+G P+ LG + L KG+W+G L G+ + ++L +T+ TDW+ + AR R+
Sbjct: 181 CYYLIGIPVGVVLGNLIHLQEKGVWIGMLFGTFVQTVMLITITLKTDWENQVVIARNRV 357
>TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120, partial
(18%)
Length = 723
Score = 85.9 bits (211), Expect = 1e-17
Identities = 38/92 (41%), Positives = 59/92 (63%)
Frame = +1
Query: 293 LCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMG 352
L VS+ S+ LSG+A G G+++ AYVNLG YY++G P+ LG KG+W+G
Sbjct: 1 LAVSILLYSVPPVLSGVALGAGWRSTVAYVNLGCYYIIGLPVGIVLGNVFHWQVKGIWIG 180
Query: 353 TLTGSILNVIILAVVTMLTDWQKEATKARERI 384
L G+++ I+L ++T TDW+++ T AR R+
Sbjct: 181 MLFGTLVQTIVLVIITYRTDWEEQVTIARNRV 276
>TC16376 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (17%)
Length = 550
Score = 84.7 bits (208), Expect = 2e-17
Identities = 40/79 (50%), Positives = 51/79 (63%)
Frame = +2
Query: 306 LSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILA 365
LSG+A G G+QT AYVN+G YY +G P LGF K A G+W+G L G++L IIL
Sbjct: 11 LSGVAVGCGWQTFVAYVNVGCYYGIGIPFGAVLGFYFKFGAMGIWLGMLAGTVLQTIILV 190
Query: 366 VVTMLTDWQKEATKARERI 384
VT TDW KE +A +R+
Sbjct: 191 WVTFRTDWNKEIEEATKRL 247
>BP069580
Length = 483
Score = 84.7 bits (208), Expect = 2e-17
Identities = 38/75 (50%), Positives = 51/75 (67%)
Frame = -1
Query: 307 SGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILAV 366
SG ARG G Q IGA++NLG+YYLVG P+A L F L + KGLW+G + I+ V L +
Sbjct: 264 SGTARGCGLQKIGAFINLGSYYLVGIPLAMVLAFVLHIGGKGLWLGIICALIVQVFSLMI 85
Query: 367 VTMLTDWQKEATKAR 381
+T+ DW+KEA + R
Sbjct: 84 ITLRIDWEKEARRPR 40
>TC9722 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (16%)
Length = 571
Score = 84.3 bits (207), Expect = 3e-17
Identities = 41/79 (51%), Positives = 52/79 (64%)
Frame = +3
Query: 306 LSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKGLWMGTLTGSILNVIILA 365
LSG+A G G QT AYVN+G YY VG P+ LGF K AKG+W+G L G+++ IIL
Sbjct: 3 LSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVMQTIILM 182
Query: 366 VVTMLTDWQKEATKARERI 384
VT TDW KE +A +R+
Sbjct: 183 WVTFRTDWIKEVEEASKRL 239
>BP042877
Length = 489
Score = 84.0 bits (206), Expect = 4e-17
Identities = 41/98 (41%), Positives = 59/98 (59%)
Frame = -1
Query: 287 TDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNA 346
+D+ P L +S+ I LSG+A G G+ AYVN+G YY VG P+ LGF + A
Sbjct: 489 SDLCPLLALSILLHGIQPVLSGVAVGCGWPAFVAYVNVGCYYGVGIPLGSVLGFYFQFGA 310
Query: 347 KGLWMGTLTGSILNVIILAVVTMLTDWQKEATKARERI 384
+G+W+G L G+ + IIL VT TDW +E +A +R+
Sbjct: 309 QGIWLGMLGGTTMPTIILMWVTFRTDWDQEVEEAAQRL 196
>TC12808 weakly similar to GB|CAA16564.1|2827556|ATT12H17 predicted protein
{Arabidopsis thaliana;}, partial (32%)
Length = 624
Score = 84.0 bits (206), Expect = 4e-17
Identities = 49/166 (29%), Positives = 84/166 (50%), Gaps = 5/166 (3%)
Frame = +1
Query: 228 STRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGNAYSNDKEVVDYVT 287
STRVSNELGA A + V++ +G+ I S + + R I G +S+D+ +++ V
Sbjct: 4 STRVSNELGANQAGRAYRSACVSLALGLILGCIGSLVMVAARGIWGPLFSHDRGIINGVK 183
Query: 288 DMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAK 347
+ + + + + GI RG +G Y NLG +Y + P+ F L L
Sbjct: 184 KTMLLMALVEVFNFPLAVCGGIVRGTARPWLGMYANLGGFYFLALPLGVVFAFKLNLGLF 363
Query: 348 GLWMGTLTGSILNVIILAVVTMLTDWQKEATKAR-----ERIAEKP 388
GL++G +TG + + +L + +W++EA+KA+ E + E P
Sbjct: 364 GLFIGLITGIVTCLSLLVIFIARINWEEEASKAQALASNEHVKEVP 501
>TC12429 similar to UP|Q9FKQ1 (Q9FKQ1) Emb|CAB89401.1 (AT5g65380/MNA5_11),
partial (22%)
Length = 523
Score = 81.3 bits (199), Expect = 3e-16
Identities = 40/101 (39%), Positives = 61/101 (59%), Gaps = 1/101 (0%)
Frame = +1
Query: 282 VVDYVTDMVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFG 341
V+D V ++ L ++ +SI LSG+A G G+Q+ AY+NLG YYLVG P+ + +G+
Sbjct: 1 VLDEVNNLSLLLAFTILLNSIQPVLSGVAVGSGWQSYVAYINLGCYYLVGVPLGFIMGWV 180
Query: 342 LKLNAKGLWMGTL-TGSILNVIILAVVTMLTDWQKEATKAR 381
G+W G + G+ L +IL V+T+ DW EA KA+
Sbjct: 181 FNQGVMGIWAGMIFGGTALQTLILGVITIRCDWDGEAEKAK 303
>AV776569
Length = 633
Score = 80.1 bits (196), Expect = 6e-16
Identities = 40/129 (31%), Positives = 70/129 (54%)
Frame = +2
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M ALETLCGQ +GA + +G Y+ + + L + +++ +L + GQ EI+
Sbjct: 236 MGSALETLCGQAFGAGQSRMLGVYMQRSWVILFTTALILVPAYVWSPPILRVIGQTTEIS 415
Query: 61 QAAREYCICLIPALFGHAVLQSLIRYFQIQSMIFPMVFSSIVILCLHVPICWCLVFKFGL 120
+AA ++ + ++P LF +A + ++ Q Q + M++ S +L LH+ W L+ K G
Sbjct: 416 EAAGKFALWMLPQLFAYAFNFPMQKFLQSQRKVQVMLWISATVLVLHIFFSWLLILKLGW 595
Query: 121 GHVGAAFAI 129
G GAA +
Sbjct: 596 GLTGAAITL 622
>TC12419
Length = 667
Score = 77.8 bits (190), Expect = 3e-15
Identities = 37/87 (42%), Positives = 51/87 (58%)
Frame = +3
Query: 289 MVPFLCVSVSADSIICALSGIARGGGFQTIGAYVNLGAYYLVGAPIAYFLGFGLKLNAKG 348
+ P L +S+ DSI LSG+ARG G+Q + AYVNL +YL+G PI+ L F L KG
Sbjct: 15 VAPLLAISILLDSIQGVLSGVARGCGWQHLAAYVNLATFYLIGLPISCLLAFKTSLQYKG 194
Query: 349 LWMGTLTGSILNVIILAVVTMLTDWQK 375
LW+G + G + L ++T W K
Sbjct: 195 LWIGLICGMVCQAGTLLLLTTRAKWAK 275
>AV778212
Length = 557
Score = 62.0 bits (149), Expect = 2e-10
Identities = 26/77 (33%), Positives = 48/77 (61%)
Frame = +1
Query: 1 MAGALETLCGQTYGAEEFSKIGNYICSAMITLILVCFPISLMWIFIDKLLLLFGQDIEIA 60
M A+ETLCGQ YGA ++ +G Y+ A I L + P++++++F ++LL+ G+ ++A
Sbjct: 325 MGSAVETLCGQAYGAHKYDMLGIYMQRAFIVLTITGIPLTVVYVFCKQILLVLGESDDVA 504
Query: 61 QAAREYCICLIPALFGH 77
A + LIP ++ +
Sbjct: 505 SAGAIFVYGLIPQIYAY 555
>AV417402
Length = 371
Score = 61.6 bits (148), Expect = 2e-10
Identities = 42/125 (33%), Positives = 60/125 (47%), Gaps = 3/125 (2%)
Frame = +3
Query: 96 MVFSSIVILCLHVPICWCLVFKFGLGHVGAAFAIGIAYWLN---VIWLGIYMKYSPACEK 152
+ + +++ + LH+PI + LV LG G A A W N VI L IY+ S +K
Sbjct: 3 LTYCAVLSILLHIPINYLLVSVLHLGIRGIALG---AVWTNFNLVISLIIYIWVSGIHKK 173
Query: 153 TKIVFSYNSLLYIAEFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLST 212
T S AIPS + CLEWW +EI+ ++ GLL N + + V + T
Sbjct: 174 TWTGISSACFKGWKALLNLAIPSCISVCLEWWWYEIMILLCGLLINPHATVASMGVLIQT 353
Query: 213 TTLHY 217
T L Y
Sbjct: 354 TALIY 368
>BP055219
Length = 527
Score = 47.8 bits (112), Expect = 3e-06
Identities = 28/108 (25%), Positives = 49/108 (44%)
Frame = -1
Query: 167 EFCQFAIPSGLMFCLEWWSFEILTIVAGLLPNSQLETSVLSVCLSTTTLHYFIPHAIGAS 226
EF + + SG+M C+E W +L ++ G L N+++ LS+C+S L IP + A+
Sbjct: 479 EFVKLSAASGVMLCVENWYCRVLIVMTGNLQNAEIAVDALSICMSINGLEMMIPLSFFAA 300
Query: 227 ASTRVSNELGAGNPRAAKGAVRVAVIIGIAEAVIVSTLFLCFRNIIGN 274
+ N A + + V+V F+C R++ N
Sbjct: 299 TG*ENIGGA*SSNQNCA-----------LLKLVVVFFFFVCCRSLTKN 189
>AV413805
Length = 418
Score = 41.2 bits (95), Expect = 3e-04
Identities = 17/21 (80%), Positives = 20/21 (94%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEFSKI 21
M+GALETLCGQT+GAEEF K+
Sbjct: 354 MSGALETLCGQTFGAEEFGKL 416
>TC10052
Length = 554
Score = 40.0 bits (92), Expect = 7e-04
Identities = 33/115 (28%), Positives = 57/115 (48%), Gaps = 10/115 (8%)
Frame = +3
Query: 101 IVILCLHVPICWCLVFK-FGLGHVGAAFAIGIAYWLNVIWLGIYMKYSPACEKTKIVFSY 159
+ +L LH+ C + K FG +GA IG + Y + + +V +
Sbjct: 6 LYLLVLHI----C*M*KYFGYNFIGALHGIGE-----------FFGYVISLVSSSLVLLH 140
Query: 160 NSLLYIAEFCQFAIPSGLM---------FCLEWWSFEILTIVAGLLPNSQLETSV 205
+L+++ C + I S ++ + LEWW FE+LT+++ LLPN +L+TSV
Sbjct: 141 LNLIFL---CYYFISSVILNHDLALFDHWSLEWWPFELLTVLS-LLPNPELKTSV 293
>TC19977 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (20%)
Length = 452
Score = 31.2 bits (69), Expect = 0.31
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +3
Query: 1 MAGALETLCGQTYGAEEF 18
M A+ETLCGQ YGA++F
Sbjct: 396 MGSAVETLCGQAYGAKKF 449
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.329 0.142 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,691,511
Number of Sequences: 28460
Number of extensions: 123979
Number of successful extensions: 1028
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1020
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1024
length of query: 396
length of database: 4,897,600
effective HSP length: 92
effective length of query: 304
effective length of database: 2,279,280
effective search space: 692901120
effective search space used: 692901120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144375.8


