

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
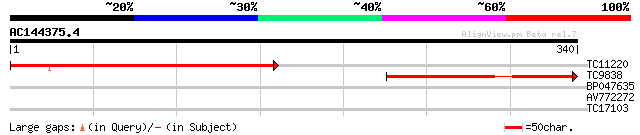
Query= AC144375.4 - phase: 0
(340 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11220 similar to UP|Q96259 (Q96259) AR401, partial (37%) 275 1e-74
TC9838 similar to UP|Q96347 (Q96347) SLL2, partial (28%) 191 2e-49
BP047635 27 3.8
AV772272 26 8.4
TC17103 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein... 26 8.4
>TC11220 similar to UP|Q96259 (Q96259) AR401, partial (37%)
Length = 536
Score = 275 bits (702), Expect = 1e-74
Identities = 138/163 (84%), Positives = 147/163 (89%), Gaps = 2/163 (1%)
Frame = +3
Query: 1 MAGIRLQPEDSDVSQQARAVAL--VDLVSDDDRSIAADSWSIKSEYGSTLDDDQRHADAA 58
MAGIRLQPEDSDVSQQ R+V+L DLVSDDDRSIAADSWSIKSEYGSTLDDDQRHADAA
Sbjct: 48 MAGIRLQPEDSDVSQQVRSVSLSTADLVSDDDRSIAADSWSIKSEYGSTLDDDQRHADAA 227
Query: 59 EALSNVNLRAASDYSSDKDEPDAEAVSSMLGFQSYWDAAYTDELTNFHEHGHAGEVWFGD 118
EALSN NL SDYSSDK+EPDAEAV+S LGFQSYWDAAY+DELTNF EHGHAGEV FG
Sbjct: 228 EALSNANLPPPSDYSSDKEEPDAEAVASRLGFQSYWDAAYSDELTNFREHGHAGEVGFGV 407
Query: 119 NVMEVVASWTKTLCIDISQGRLPNHVDDVKADAGELDDKLLSS 161
+VMEVVASWT+ LCIDISQGRLPNH DDVKA+A EL DK+LSS
Sbjct: 408 DVMEVVASWTRALCIDISQGRLPNHTDDVKAEASELGDKILSS 536
>TC9838 similar to UP|Q96347 (Q96347) SLL2, partial (28%)
Length = 562
Score = 191 bits (484), Expect = 2e-49
Identities = 95/114 (83%), Positives = 101/114 (88%)
Frame = +1
Query: 227 FQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTKDELVQEVESFN 286
FQLV+DKGTLDAIGLHPDG VKRMMYWDSV++LVAPGGILV+TSCN+TKDELVQEVESFN
Sbjct: 1 FQLVVDKGTLDAIGLHPDGSVKRMMYWDSVAKLVAPGGILVITSCNNTKDELVQEVESFN 180
Query: 287 QRKSATAPEPEVAKDDESCRDPLFQYVSHVRTYPTFMFGGSVGSRVATVAFLRK 340
QR A +ESCRDPLFQYVSHVRTYPTFMFGGSVGSRVATVAFLRK
Sbjct: 181 QRNHA----------EESCRDPLFQYVSHVRTYPTFMFGGSVGSRVATVAFLRK 312
>BP047635
Length = 467
Score = 27.3 bits (59), Expect = 3.8
Identities = 13/51 (25%), Positives = 23/51 (44%), Gaps = 3/51 (5%)
Frame = +2
Query: 76 KDEPDAEAVSSMLGFQSYW---DAAYTDELTNFHEHGHAGEVWFGDNVMEV 123
K P+A+ ++ W + YT+ T++H H H W+ + EV
Sbjct: 83 KTSPNAQIKIHNFNNRTVWVVREGHYTNLQTHYHHHHHRDSAWWVGGLAEV 235
>AV772272
Length = 436
Score = 26.2 bits (56), Expect = 8.4
Identities = 14/36 (38%), Positives = 22/36 (60%)
Frame = -1
Query: 234 GTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVT 269
GTLD + +H D PV R++ SVS + I+V++
Sbjct: 109 GTLDNVYVHLDSPV*RIIK*SSVSNIKKNYQIVVIS 2
>TC17103 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (19%)
Length = 685
Score = 26.2 bits (56), Expect = 8.4
Identities = 14/31 (45%), Positives = 20/31 (64%)
Frame = +2
Query: 203 ANRDGFPNIKFLVDDVLETKLEQVFQLVMDK 233
A++D PNIKF V VLE+ + Q V++K
Sbjct: 146 ASKDRVPNIKFNVAKVLESIFPILDQSVVEK 238
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,195,919
Number of Sequences: 28460
Number of extensions: 63795
Number of successful extensions: 181
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 179
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 179
length of query: 340
length of database: 4,897,600
effective HSP length: 91
effective length of query: 249
effective length of database: 2,307,740
effective search space: 574627260
effective search space used: 574627260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC144375.4


