

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.3 + phase: 0
(368 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
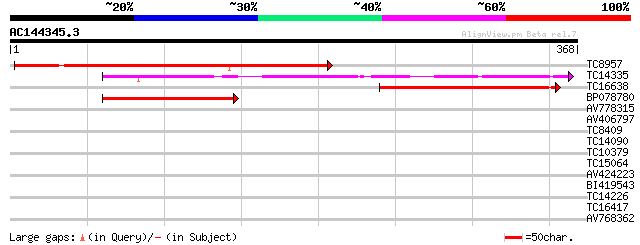
Score E
Sequences producing significant alignments: (bits) Value
TC8957 weakly similar to UP|Q9ZUJ5 (Q9ZUJ5) T2K10.2 protein, par... 248 1e-66
TC14335 similar to UP|MEP1_SOYBN (P29136) Metalloendoproteinase ... 166 4e-42
TC16638 weakly similar to UP|MEP1_SOYBN (P29136) Metalloendoprot... 166 4e-42
BP078780 94 5e-20
AV778315 29 1.1
AV406797 28 2.4
TC8409 similar to UP|Q9FPS8 (Q9FPS8) Ubiquitin-specific protease... 28 3.2
TC14090 homologue to UP|ALF_CICAR (O65735) Fructose-bisphosphate... 28 3.2
TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast ... 27 5.4
TC15064 similar to UP|Q948P0 (Q948P0) Aspartic proteinase alpha,... 27 5.4
AV424223 27 7.0
BI419543 27 7.0
TC14226 UP|Q9MBE4 (Q9MBE4) Cytochrome P450, complete 26 9.2
TC16417 homologue to UP|AAR83884 (AAR83884) Ly200 protein, complete 26 9.2
AV768362 26 9.2
>TC8957 weakly similar to UP|Q9ZUJ5 (Q9ZUJ5) T2K10.2 protein, partial (20%)
Length = 724
Score = 248 bits (633), Expect = 1e-66
Identities = 120/214 (56%), Positives = 151/214 (70%), Gaps = 8/214 (3%)
Frame = +1
Query: 4 QNQRYYYHIFYITLISITLTSVSARFFPDPSSMPAWNSSNAPP-GAWDGYKNFTGCSRGK 62
Q + + + + L S VSAR FPD +P++ PP GAWD ++NF+GC RG+
Sbjct: 91 QQPQLHIILTLVHLFSFGTAVVSARLFPD---VPSFIPDGVPPSGAWDAFRNFSGCHRGE 261
Query: 63 TYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNY 122
YDGLS LK+YF FGYIP+ PPSNF+DDFD+ALE+A++TYQKNFNLNITG+LDD T+
Sbjct: 262 NYDGLSNLKSYFKRFGYIPHAPPSNFSDDFDDALEAAIKTYQKNFNLNITGDLDDKTLRQ 441
Query: 123 IVKPRCGVADIINGTTSMN-------SGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLT 175
I+ PRCGVADIINGTT+MN + S+ +FHTV+HY+ FPGQPR P G Q LT
Sbjct: 442 IMLPRCGVADIINGTTTMNAAGNQDETASHGDSNLHFHTVSHYALFPGQPRGPAGKQELT 621
Query: 176 YAFDPSENLDDATKQVFANAFNQWSKVTTITFTE 209
YAF P +L DA K VF AF +WS+VTT+TF E
Sbjct: 622 YAFYPGNSLSDAVKSVFTGAFARWSEVTTLTFRE 723
>TC14335 similar to UP|MEP1_SOYBN (P29136) Metalloendoproteinase 1 precursor
(SMEP1) , partial (9%)
Length = 1574
Score = 166 bits (421), Expect = 4e-42
Identities = 110/309 (35%), Positives = 160/309 (51%), Gaps = 3/309 (0%)
Frame = +3
Query: 61 GKTYDGLSKLKNYFNHFGYIPN---GPPSNFTDDFDEALESAVRTYQKNFNLNITGELDD 117
G + GL +K Y + GYI + SN TD F E L+ A+ YQKNFNL +TG +D
Sbjct: 249 GDSVKGLENIKKYLDALGYIKDLVLALNSNDTDKFTEDLKPAIMEYQKNFNLPVTGVIDQ 428
Query: 118 ATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYA 177
N +++PRCGV D T++N G + +P W + L+YA
Sbjct: 429 NLYNTLIQPRCGVPDF----TTLNVGTTPAF---------------KPWWKSEKKELSYA 551
Query: 178 FDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVL 237
F P N+ D + VF AF++WS T++ FTE ++ +SDIKI F D G G G +
Sbjct: 552 FHPQNNVSDDIRTVFREAFDRWSNETSLNFTETATFDASDIKIAFVKWD-GKG----GRV 716
Query: 238 GTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEE 297
G +S G + LD E +V L +VV+HEIGHLLGLGHS +E
Sbjct: 717 GAAYTNYSMHAGGMFLDIDEQYV---------------LLNVVMHEIGHLLGLGHSYEKE 851
Query: 298 AIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLFLS 357
+IMYP + +K++ DD + IQ + G++ N + +++ DSS GG H S++
Sbjct: 852 SIMYPIV--MREKIDFTDDDRKRIQKVTGNSNNGGTSASSSGTTTDSSSGGGHD-SVWQR 1022
Query: 358 LVFLGFGFL 366
+ LGF F+
Sbjct: 1023NLILGFAFM 1049
>TC16638 weakly similar to UP|MEP1_SOYBN (P29136) Metalloendoproteinase 1
precursor (SMEP1) , partial (29%)
Length = 621
Score = 166 bits (421), Expect = 4e-42
Identities = 84/118 (71%), Positives = 101/118 (85%), Gaps = 1/118 (0%)
Frame = +2
Query: 241 AHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIM 300
AHAFSPT+GR HLD AEDWVV+GDV++S+L+ AVDLESV VHEIGHLLGLGHSS+EEAIM
Sbjct: 2 AHAFSPTNGRFHLDAAEDWVVSGDVSKSALATAVDLESVAVHEIGHLLGLGHSSVEEAIM 181
Query: 301 YPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTAT-SRDRDSSFGGRHGVSLFLS 357
+PTISSRTKKV L DDI+GIQ LYG+NP+F G++ T S +RD+S GGR +S FL+
Sbjct: 182 FPTISSRTKKVVLAEDDIKGIQYLYGTNPSFNGSSATTFSPERDASDGGRR-LSCFLA 352
>BP078780
Length = 516
Score = 93.6 bits (231), Expect = 5e-20
Identities = 42/88 (47%), Positives = 65/88 (73%)
Frame = +3
Query: 61 GKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATM 120
G DGLS++++Y + FGY+ + SNFT+++ E +SA+ +QKN+NL +TG+LD+
Sbjct: 246 GDIVDGLSQVQDYLDLFGYLNSTLHSNFTNEYTENAKSAIEEFQKNYNLQVTGQLDNNLY 425
Query: 121 NYIVKPRCGVADIINGTTSMNSGKFNSS 148
+ + +PRCGV DIINGTT+MNS K N++
Sbjct: 426 HVLSQPRCGVPDIINGTTTMNSRKANTT 509
>AV778315
Length = 559
Score = 29.3 bits (64), Expect = 1.1
Identities = 16/56 (28%), Positives = 26/56 (45%)
Frame = -3
Query: 158 YSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSY 213
+ FPG+P W E ++ + + LD ++++ AFN TIT E Y
Sbjct: 539 HGHFPGKPDWSETSRFVAFTM-----LDSVKREIYI-AFNTSHLPVTITLPERAGY 390
>AV406797
Length = 418
Score = 28.1 bits (61), Expect = 2.4
Identities = 16/33 (48%), Positives = 18/33 (54%)
Frame = +1
Query: 14 YITLISITLTSVSARFFPDPSSMPAWNSSNAPP 46
Y +S T S S R P PS P+ NSSN PP
Sbjct: 229 YSRSVSWTDGSPSRRKPPPPSQPPSSNSSNKPP 327
>TC8409 similar to UP|Q9FPS8 (Q9FPS8) Ubiquitin-specific protease 16,
partial (4%)
Length = 709
Score = 27.7 bits (60), Expect = 3.2
Identities = 26/101 (25%), Positives = 43/101 (41%), Gaps = 6/101 (5%)
Frame = +1
Query: 181 SENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTL 240
S ++DD + +F ++ WS + + ++ +S SSS + F D + +D V
Sbjct: 136 STSIDDFSDYIFGDSGRGWSSLWRNSDSDTSSTSSS--PLNFRHSPLSDMDRYDSVSPVA 309
Query: 241 AHAFSPT------DGRLHLDKAEDWVVNGDVTESSLSNAVD 275
PT DG H + D V V +SSL +D
Sbjct: 310 TGLRIPTGSSSEIDGPSHRSREMD-VERKGVGDSSLHRKLD 429
>TC14090 homologue to UP|ALF_CICAR (O65735) Fructose-bisphosphate aldolase,
cytoplasmic isozyme , partial (98%)
Length = 1491
Score = 27.7 bits (60), Expect = 3.2
Identities = 16/53 (30%), Positives = 26/53 (48%)
Frame = -3
Query: 134 INGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDD 186
++G S++SGKFNS+ N T + SF + + TL F ++ D
Sbjct: 499 LSGGLSISSGKFNSTLVNLDTGENTSFLQNINKGLACSSTLVQGFLKEDHSTD 341
>TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast nucleoid
DNA binding protein (CND41), partial (43%)
Length = 1002
Score = 26.9 bits (58), Expect = 5.4
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = -3
Query: 30 FPDPSSMPAWNSSNAPPGAWDGYKNF 55
FPDP +P + S APP W Y +F
Sbjct: 427 FPDPIRLPDFTGSVAPPD-WPEYDSF 353
>TC15064 similar to UP|Q948P0 (Q948P0) Aspartic proteinase alpha, partial
(77%)
Length = 1444
Score = 26.9 bits (58), Expect = 5.4
Identities = 17/51 (33%), Positives = 25/51 (48%)
Frame = -1
Query: 86 SNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADIING 136
++F+ FDE L T NFN+ +T E D + + RC + DI G
Sbjct: 202 ASFSGSFDEILMLDNST--TNFNMILTEETRDCSRSIGYLARCSIFDICFG 56
>AV424223
Length = 288
Score = 26.6 bits (57), Expect = 7.0
Identities = 22/70 (31%), Positives = 27/70 (38%), Gaps = 3/70 (4%)
Frame = +1
Query: 287 LLGLGHSSIEEAIMYPTISSRTKKVELESDDI---EGIQMLYGSNPNFTGTTTATSRDRD 343
L L HSS + SSR KK EL DD+ G G + T + DRD
Sbjct: 19 LPALNHSSEQTIGKEDIWSSRVKKRELFLDDVGGTHGTSSAAGVGSSLTSSAKGKRSDRD 198
Query: 344 SSFGGRHGVS 353
R +S
Sbjct: 199 GKGHSREVLS 228
>BI419543
Length = 585
Score = 26.6 bits (57), Expect = 7.0
Identities = 17/37 (45%), Positives = 21/37 (55%), Gaps = 3/37 (8%)
Frame = +2
Query: 60 RGKTYDGLSKLKNYFN--HFGYIPNG-PPSNFTDDFD 93
R +T DGL LKNY + ++G I G PP F FD
Sbjct: 338 RTETDDGLLPLKNYLDAQYYGEIAIGTPPQTFDVIFD 448
>TC14226 UP|Q9MBE4 (Q9MBE4) Cytochrome P450, complete
Length = 1878
Score = 26.2 bits (56), Expect = 9.2
Identities = 12/33 (36%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Frame = +3
Query: 9 YYHIFYITLISITLTSVSARFF----PDPSSMP 37
YY +FY+ L +I + +R F P P+S+P
Sbjct: 63 YYSLFYVALFAIIKLFLGSRKFKNLPPGPTSLP 161
>TC16417 homologue to UP|AAR83884 (AAR83884) Ly200 protein, complete
Length = 719
Score = 26.2 bits (56), Expect = 9.2
Identities = 12/25 (48%), Positives = 13/25 (52%)
Frame = +3
Query: 344 SSFGGRHGVSLFLSLVFLGFGFLSL 368
S FG HG F LVF G F+ L
Sbjct: 483 SQFGYGHGTKSFSKLVFFGLWFVDL 557
>AV768362
Length = 541
Score = 26.2 bits (56), Expect = 9.2
Identities = 13/45 (28%), Positives = 19/45 (41%), Gaps = 4/45 (8%)
Frame = -3
Query: 18 ISITLTSVSARFFPDPSSMPAWNSSNAPPGAWD----GYKNFTGC 58
+ I +S+ +PS W + P G WD G +NF C
Sbjct: 323 MKIKSSSLCLASHEEPSQCSCWKDESTPKGDWDNRFLGAQNFFLC 189
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,435,558
Number of Sequences: 28460
Number of extensions: 90412
Number of successful extensions: 549
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 543
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 545
length of query: 368
length of database: 4,897,600
effective HSP length: 92
effective length of query: 276
effective length of database: 2,279,280
effective search space: 629081280
effective search space used: 629081280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144345.3


