

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.10 + phase: 0
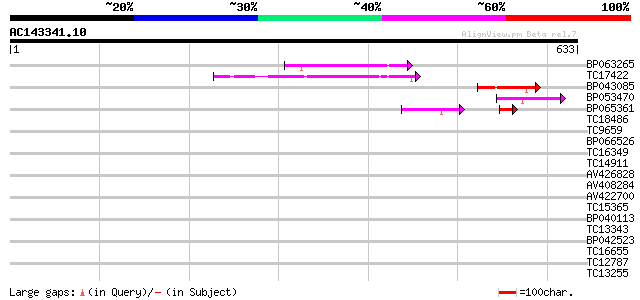
(633 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP063265 86 2e-17
TC17422 similar to GB|CAB10335.1|2244913|ATFCA4 SEN1 like protei... 80 1e-15
BP043085 65 4e-11
BP053470 56 2e-08
BP065361 41 6e-07
TC18486 similar to UP|Q9LZQ9 (Q9LZQ9) ATP-dependent RNA helicase... 36 0.016
TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATP... 33 0.11
BP066526 32 0.24
TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulat... 32 0.31
TC14911 homologue to UP|Q93X55 (Q93X55) Peroxin 6 (Fragment), pa... 32 0.40
AV426828 31 0.53
AV408284 31 0.53
AV422700 30 1.5
TC15365 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {... 29 2.6
BP040113 29 2.6
TC13343 similar to UP|Q9FMR9 (Q9FMR9) Ruv DNA-helicase-like prot... 29 2.6
BP042523 29 2.6
TC16655 homologue to UP|O99018 (O99018) Chloroplast protease pre... 28 3.4
TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (T... 28 4.5
TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%) 28 4.5
>BP063265
Length = 497
Score = 85.9 bits (211), Expect = 2e-17
Identities = 51/147 (34%), Positives = 81/147 (54%), Gaps = 5/147 (3%)
Frame = +2
Query: 308 REIQKELRTLSREERKR----QQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDE 363
++++ E LS + K+ ++ ++ +++DVI T +GA +L N F V+IDE
Sbjct: 29 QQLKDEQGELSSSDEKKYKALKRATEREISQSADVICCTCVGAGDPRLANFRFRQVLIDE 208
Query: 364 AAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVT 422
+ QA E C IPL+ G + +L GDH QL P I +A + GL ++LFERL L +
Sbjct: 209 STQATEPECLIPLVLGAKQVVLVGDHCQLGPVIMCKKAARAGLAQSLFERLVLLGVKPI- 385
Query: 423 SMLTVQYRMHQLIMDWSSKELYNSKVK 449
L VQYRMH + ++ S Y ++
Sbjct: 386 -RLQVQYRMHPCLSEFPSNSFYEGTLQ 463
>TC17422 similar to GB|CAB10335.1|2244913|ATFCA4 SEN1 like protein
{Arabidopsis thaliana;}, partial (37%)
Length = 868
Score = 80.1 bits (196), Expect = 1e-15
Identities = 78/241 (32%), Positives = 111/241 (45%), Gaps = 10/241 (4%)
Frame = +3
Query: 228 KILACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDI 287
++L CA SN A+D IV R+ R V + P +V L A
Sbjct: 6 RVLVCAPSNSALDEIVLRV---RNAGVHDENGRGYCPNIVRIGLKAH------------- 137
Query: 288 RKEMKVLN-GKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGA 346
+K ++ +LLK K + + + T A ++ + ++ +TL +
Sbjct: 138 -HSIKAVSLDELLKQKRSSGNKSSTDKQSTGPAGSNDDTLRAA--ILDEATIVFSTLSFS 308
Query: 347 SSKKLG--NTSFDLVIIDEAAQALEVACWIPLLKGTRCI-LAGDHLQLPPTIQSVEAEKK 403
S L + SFD+VIIDEAAQA+E A +PL + + L GD QLP T+ S A+
Sbjct: 309 GSNSLSKLSRSFDVVIIDEAAQAVEPAVLVPLANQCKKVFLVGDPAQLPATVISDIAKNH 488
Query: 404 GLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSK------VKAHACVASH 457
G G +LFERL +L G V ML QYRMH I + S+E Y +K H A H
Sbjct: 489 GYGTSLFERL-KLAGYPV-KMLKTQYRMHPEIRSFPSREFYEDSLQDGDGIKLHTTRAWH 662
Query: 458 M 458
M
Sbjct: 663 M 665
>BP043085
Length = 466
Score = 64.7 bits (156), Expect = 4e-11
Identities = 39/77 (50%), Positives = 49/77 (62%), Gaps = 7/77 (9%)
Frame = -2
Query: 523 IITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKE------- 575
IITPY QV+ +K E + DI++ +V+ FQG+EKE IIIS VRS K
Sbjct: 465 IITPYKQQVLKIKQTLENEG-MPDIKVGSVEQFQGQEKEVIIISTVRSTIKHNEIDRVHC 289
Query: 576 VGFLSDRRRMNVAVTRA 592
+GFLS+ RR NVAVTRA
Sbjct: 288 LGFLSNFRRFNVAVTRA 238
>BP053470
Length = 498
Score = 56.2 bits (134), Expect = 2e-08
Identities = 31/80 (38%), Positives = 48/80 (59%), Gaps = 3/80 (3%)
Frame = -2
Query: 544 LKDIEISTVDGFQGREKEAIIISMVRSN---SKKEVGFLSDRRRMNVAVTRARRQCCIVC 600
+ +EI T+D +QGR+K+ I++S VRS+ + L D R+NVA+TRA+R+ +V
Sbjct: 491 ITSLEIHTIDXYQGRDKDCILVSFVRSSENPTSYAASLLGDWHRINVALTRAKRKLIMVG 312
Query: 601 DTETVSSDGFLKRLIEYFEE 620
T+ LK LI+ EE
Sbjct: 311 SRRTLLKVPLLKLLIKKVEE 252
>BP065361
Length = 442
Score = 40.8 bits (94), Expect(2) = 6e-07
Identities = 27/78 (34%), Positives = 39/78 (49%), Gaps = 8/78 (10%)
Frame = +3
Query: 438 WSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDT------AGCDMEEKKDE 491
W+SKE+Y +K+ V SH+L D V+ T T+ LLL+DT D E D
Sbjct: 33 WASKEMYGGLLKSFKTVFSHLLVDFPFVQPTWITQCPLLLLDTRMPYESMSVDCGEHLDP 212
Query: 492 --EDSTLNEGESEVAMAH 507
S NE E+++ + H
Sbjct: 213 AGTGSFYNE*EADIVLQH 266
Score = 29.6 bits (65), Expect(2) = 6e-07
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +2
Query: 548 EISTVDGFQGREKEAIIISM 567
E+ST+D FQG E + +I+SM
Sbjct: 293 EVSTIDSFQGLEADVVILSM 352
>TC18486 similar to UP|Q9LZQ9 (Q9LZQ9) ATP-dependent RNA helicase-like
protein, partial (22%)
Length = 549
Score = 36.2 bits (82), Expect = 0.016
Identities = 28/103 (27%), Positives = 43/103 (41%), Gaps = 6/103 (5%)
Frame = +1
Query: 187 QKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEV------KRGSKILACAASNIAVD 240
QK+ KAL +L G G+GKTT + + +L+ V +RG ++AC
Sbjct: 193 QKEEFFKALRENQTLILVGETGSGKTTQIPQFVLEAVDLESPDRRGKMMVACTQPRRVAA 372
Query: 241 NIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGL 283
V R V + V IG + D + VL+ G+
Sbjct: 373 MSVSRRVAEEMD-VTIGEEVGYSIRFEDCSSSKTVLKYLTDGM 498
>TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (74%)
Length = 1060
Score = 33.5 bits (75), Expect = 0.11
Identities = 40/166 (24%), Positives = 72/166 (43%), Gaps = 9/166 (5%)
Frame = +1
Query: 201 FLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVD--NIVERLVPHRVKLVRIGH 258
FLL+GPPGTGK+ + + S + ++S++ E+LV + ++ R
Sbjct: 571 FLLYGPPGTGKSYLAKAV---ATEADSTFFSVSSSDLGSKWMGESEKLVSNLFQMARESA 741
Query: 259 PARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVL-------NGKLLKTKEKNTRREIQ 311
P+ + +DS L Q G+ S + I+ E+ V + K+L NT +
Sbjct: 742 PSIIFVDEIDS-LCGQRGEGNESEASRRIKTELLVQMQGVGNNDQKVLVLAATNTPYALD 918
Query: 312 KELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFD 357
+ +R R KR + + DV K + +G + L + F+
Sbjct: 919 QAIR---RRFDKRIYIPLPDV-KARQHMFKVHLGDTPHNLAESDFE 1044
>BP066526
Length = 440
Score = 32.3 bits (72), Expect = 0.24
Identities = 17/45 (37%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Frame = -2
Query: 205 GPPGTGKTTTVVEI--ILQEVKRGSKILACAASNIAVDNIVERLV 247
GPPGTGKT T V+I +L + L SN A++++ E+++
Sbjct: 436 GPPGTGKTDTAVQILNVLYHNCPSQRTLIITHSNQALNDLFEKIM 302
>TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulatory subunit
6B homolog, partial (65%)
Length = 901
Score = 32.0 bits (71), Expect = 0.31
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 11/98 (11%)
Frame = +3
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDY 185
L K + V HR +AL+ V A I +L +P V+ D+ I K
Sbjct: 372 LLKPSASVALHRHSNALVD----VLPPEADSSISLLSQSEKPDVTYNDIGGCDIQKQ--- 530
Query: 186 SQKDAISKALSSKNVF-----------LLHGPPGTGKT 212
++A+ L+ ++ LL+GPPGTGKT
Sbjct: 531 EIREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKT 644
>TC14911 homologue to UP|Q93X55 (Q93X55) Peroxin 6 (Fragment), partial (32%)
Length = 1418
Score = 31.6 bits (70), Expect = 0.40
Identities = 13/25 (52%), Positives = 17/25 (68%)
Frame = +2
Query: 188 KDAISKALSSKNVFLLHGPPGTGKT 212
KD + L ++ LL+GPPGTGKT
Sbjct: 293 KDLFASGLRKRSGVLLYGPPGTGKT 367
>AV426828
Length = 401
Score = 31.2 bits (69), Expect = 0.53
Identities = 11/17 (64%), Positives = 16/17 (93%)
Frame = +1
Query: 202 LLHGPPGTGKTTTVVEI 218
LL+GPPGTGKT+T++ +
Sbjct: 121 LLYGPPGTGKTSTILAV 171
>AV408284
Length = 425
Score = 31.2 bits (69), Expect = 0.53
Identities = 39/145 (26%), Positives = 59/145 (39%), Gaps = 7/145 (4%)
Frame = +1
Query: 170 SKKDVVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKI 229
S K+ +F + L + K L + LL+GPPGTGKT I +K
Sbjct: 7 SIKEALFELVILPLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAI--------AKE 162
Query: 230 LACAASNIAVDNIVERLVPHRVKLV----RIGH---PARLLPQVVDSALDAQVLRGDNSG 282
N+ + N++ + KLV + H PA + VDS L Q D+
Sbjct: 163 SGAVFINVRISNLMSKWFGDAQKLVAAVFSLAHKLQPAIIFIDEVDSFL-GQRRTTDHEA 339
Query: 283 LANDIRKEMKVLNGKLLKTKEKNTR 307
L N + M + +G T ++N R
Sbjct: 340 LLNMKTEFMALWDG---FTTDQNAR 405
>AV422700
Length = 472
Score = 29.6 bits (65), Expect = 1.5
Identities = 21/62 (33%), Positives = 28/62 (44%)
Frame = +2
Query: 202 LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIGHPAR 261
LL+GPPGTGK T++V ++QE C A + PH V G R
Sbjct: 320 LLYGPPGTGK-TSLVRAVVQE---------CGAHLTIIS-------PHSVHRAHAGESER 448
Query: 262 LL 263
+L
Sbjct: 449 IL 454
>TC15365 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {Arabidopsis
thaliana;}, partial (87%)
Length = 1385
Score = 28.9 bits (63), Expect = 2.6
Identities = 25/101 (24%), Positives = 45/101 (43%), Gaps = 7/101 (6%)
Frame = +3
Query: 202 LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVE-------RLVPHRVKLV 254
LL+GPPGTGKT ++ + V + C ++ +V+ R+V +
Sbjct: 441 LLYGPPGTGKT-----LLARAVAHHTD---CTFIRVSGSELVQKYIGEGSRMVRELFVMA 596
Query: 255 RIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLN 295
R P+ + +DS A++ G G + R +++LN
Sbjct: 597 REHAPSIIFMDEIDSIGSARMESGSGXGDSEVQRTMLELLN 719
>BP040113
Length = 439
Score = 28.9 bits (63), Expect = 2.6
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +2
Query: 189 DAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILAC 232
D I + + LL GPPGTGKT + I QE+ G+K+ C
Sbjct: 251 DMIRQKKMAGRALLLAGPPGTGKTALALG-ICQEL--GTKVPFC 373
>TC13343 similar to UP|Q9FMR9 (Q9FMR9) Ruv DNA-helicase-like protein,
partial (21%)
Length = 518
Score = 28.9 bits (63), Expect = 2.6
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +1
Query: 189 DAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILAC 232
D I + + LL GPPGTGKT + I QE+ G+K+ C
Sbjct: 331 DMIRQKKMAGRALLLAGPPGTGKTALALG-ICQEL--GTKVPFC 453
>BP042523
Length = 479
Score = 28.9 bits (63), Expect = 2.6
Identities = 10/17 (58%), Positives = 15/17 (87%)
Frame = +1
Query: 202 LLHGPPGTGKTTTVVEI 218
+L GPPGTGKTT+++ +
Sbjct: 184 ILSGPPGTGKTTSILAL 234
>TC16655 homologue to UP|O99018 (O99018) Chloroplast protease precursor,
partial (39%)
Length = 934
Score = 28.5 bits (62), Expect = 3.4
Identities = 35/147 (23%), Positives = 59/147 (39%), Gaps = 18/147 (12%)
Frame = +3
Query: 84 KLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLRLEKVANEVTYHRMKDALI 143
+L + + P L Q ++ + ++ +I A + ED + L + N + L
Sbjct: 366 RLQRVKVQLPGLNQELLQKFREKNIDFAAHNAQED--SGSLLFNLIGNLAFPIILIGGLF 539
Query: 144 QLSK--GVHKGPASDLIPVLFGERQPT--------VSKKDVV--------FTSINKNLDY 185
L++ G GP P+ FG+ + V+ DV F + + L
Sbjct: 540 LLTRRPGGMGGPGGPGFPLTFGQSKAKFQMEPNTGVTFDDVAGVDEAKQDFMEVVEFLKK 719
Query: 186 SQKDAISKALSSKNVFLLHGPPGTGKT 212
++ A K V L+ GPPGTGKT
Sbjct: 720 PERFTAVGARIPKGVLLI-GPPGTGKT 797
>TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (TAT-binding
homolog 4), partial (20%)
Length = 389
Score = 28.1 bits (61), Expect = 4.5
Identities = 24/91 (26%), Positives = 41/91 (44%), Gaps = 3/91 (3%)
Frame = +3
Query: 202 LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVR--IGHP 259
LL GPPGTGKT + + G+ + NI++ +I + K V+
Sbjct: 93 LLFGPPGTGKTMLAKAV---ATEAGANFI-----NISMSSITSKWFGEGEKYVKAVFSLA 248
Query: 260 ARLLPQVV-DSALDAQVLRGDNSGLANDIRK 289
+++ P V+ +D + R +N G +RK
Sbjct: 249 SKISPSVIFVDEVDXMLXRRENPGEHEAMRK 341
>TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%)
Length = 532
Score = 28.1 bits (61), Expect = 4.5
Identities = 28/116 (24%), Positives = 48/116 (41%), Gaps = 7/116 (6%)
Frame = +1
Query: 195 LSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV-------ERLV 247
LS LL GPPGTGK T + + + E K NI+ ++V E+LV
Sbjct: 25 LSPWKGILLFGPPGTGK-TMLAKAVATECK-------TTFFNISASSVVSKWRGDSEKLV 180
Query: 248 PHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
+L R P+ + +D+ + + + ++ E+ + L +T E
Sbjct: 181 KVLFQLARHHAPSTIFLDEIDAIISQRGEARSEHEASRRLKTELLIQMDGLTRTDE 348
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,153,592
Number of Sequences: 28460
Number of extensions: 88482
Number of successful extensions: 507
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 501
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 501
length of query: 633
length of database: 4,897,600
effective HSP length: 96
effective length of query: 537
effective length of database: 2,165,440
effective search space: 1162841280
effective search space used: 1162841280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC143341.10


