

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143338.3 - phase: 0
(446 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
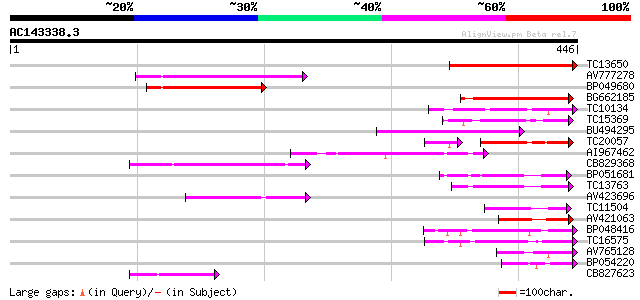
Score E
Sequences producing significant alignments: (bits) Value
TC13650 similar to UP|Q9SSL4 (Q9SSL4) F3N23.34 protein, partial ... 182 1e-46
AV777278 115 1e-26
BP049680 102 1e-22
BG662185 86 2e-17
TC10134 weakly similar to UP|O80872 (O80872) At2g30010 protein (... 80 5e-16
TC15369 similar to UP|Q9FFZ5 (Q9FFZ5) Gb|AAF00669.1, partial (17%) 78 3e-15
BU494295 78 3e-15
TC20057 similar to UP|O80872 (O80872) At2g30010 protein (At2g300... 67 5e-15
AI967462 75 2e-14
CB829368 75 3e-14
BP051681 74 4e-14
TC13763 similar to UP|O80855 (O80855) At2g30900 protein, partial... 72 2e-13
AV423696 67 8e-12
TC11504 similar to UP|O82273 (O82273) At2g31110 protein, partial... 63 9e-11
AV421063 61 4e-10
BP048416 56 1e-08
TC16575 homologue to GB|AAB61094.1|2194119|F20P5 F20P5.5 {Arabid... 56 1e-08
AV765128 55 2e-08
BP054220 54 5e-08
CB827623 54 5e-08
>TC13650 similar to UP|Q9SSL4 (Q9SSL4) F3N23.34 protein, partial (8%)
Length = 547
Score = 182 bits (461), Expect = 1e-46
Identities = 83/100 (83%), Positives = 87/100 (87%)
Frame = +2
Query: 347 GNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFR 406
GNCY EK PI+ E YWGSGSDLPTM SVE ILSSL SKVSVLN+TQ+SEYRKDGHPSIFR
Sbjct: 2 GNCYDEKDPINFEGYWGSGSDLPTMRSVEKILSSLKSKVSVLNITQMSEYRKDGHPSIFR 181
Query: 407 KFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLFHFL 446
+FWEPLRPE LSNP SYSDCI WCLPGVPD WNELLF L
Sbjct: 182 QFWEPLRPEPLSNPPSYSDCIPWCLPGVPDVWNELLFPLL 301
>AV777278
Length = 582
Score = 115 bits (289), Expect = 1e-26
Identities = 55/135 (40%), Positives = 79/135 (57%)
Frame = +3
Query: 100 NRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDC 159
+R+++ C+ F GKWV+D SYPLYN S CP++ Q C K+GR D YQ +RWQP C
Sbjct: 177 SRKLAGGRCNWFRGKWVYDR-SYPLYNPSTCPFIDPQFNCQKYGRPDTLYQQYRWQPFTC 353
Query: 160 DLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRA 219
L R+N + K R K++MFVGDSL+ Q+ S+ C++ S +P + S S +T
Sbjct: 354 PLPRFNAWDFLLKYRNKKIMFVGDSLSLNQFNSLACMIHSWVPNTRTSFSKQNVITTITF 533
Query: 220 EEYNATVEFLWAPLL 234
E+Y + P L
Sbjct: 534 EDYGLQLFLYRTPYL 578
>BP049680
Length = 474
Score = 102 bits (255), Expect = 1e-22
Identities = 43/95 (45%), Positives = 63/95 (66%)
Frame = +1
Query: 108 CDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVK 167
C+++ G WV+D SYP+Y+ S CP++ Q C GR D Y +RWQP CDL R++ K
Sbjct: 118 CNIYEGSWVYDE-SYPIYDYSSCPHIESQFDCLGRGRPDRKYLKYRWQPSGCDLPRFDGK 294
Query: 168 EMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIP 202
+ K +GK+LMF+GDS++ QW S+ C+L S +P
Sbjct: 295 KFLTKYKGKQLMFIGDSVSVNQWQSLKCMLHSAVP 399
>BG662185
Length = 383
Score = 85.5 bits (210), Expect = 2e-17
Identities = 39/90 (43%), Positives = 58/90 (64%), Gaps = 1/90 (1%)
Frame = +1
Query: 355 PIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFR-KFWEPLR 413
P+DV G+D N+ S+ V +N+T LSE+RKD H S++ + + L
Sbjct: 46 PLDV------GTDRRLFVIANNLTHSMKVPVYFINITTLSEFRKDAHTSVYTIRQGKMLT 207
Query: 414 PEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
PEQ ++P++Y+DCIHWCLPG+PDTWNE ++
Sbjct: 208 PEQQADPATYADCIHWCLPGLPDTWNEFIY 297
>TC10134 weakly similar to UP|O80872 (O80872) At2g30010 protein
(At2g30010/F23F1.7), partial (12%)
Length = 555
Score = 80.5 bits (197), Expect = 5e-16
Identities = 46/119 (38%), Positives = 64/119 (53%), Gaps = 2/119 (1%)
Frame = +1
Query: 330 TMSPTHLWSREWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLN 389
+MSP H W CY +K PI S+ L + V L + +V + +
Sbjct: 1 SMSPRHNRLNGWK------CYNQKHPIQFFSHLHVPEPLVVLRGV---LKRMRFQVYLQD 153
Query: 390 VTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSS--YSDCIHWCLPGVPDTWNELLFHFL 446
VT L+ +R+DGHPS++RK P+ E+ P + SDC HWCLPGVPD WNE+L +L
Sbjct: 154 VTTLTAFRRDGHPSVYRK---PISIEKSQKPGTGLSSDCSHWCLPGVPDIWNEMLSAWL 321
>TC15369 similar to UP|Q9FFZ5 (Q9FFZ5) Gb|AAF00669.1, partial (17%)
Length = 1365
Score = 77.8 bits (190), Expect = 3e-15
Identities = 48/109 (44%), Positives = 59/109 (54%), Gaps = 6/109 (5%)
Frame = -3
Query: 341 WNPESEGNCYGEKKP------IDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLS 394
WN G C EK+P + E Y+ + +S V + + KV LN+T LS
Sbjct: 958 WN--EGGECDMEKEPEKDHTKLATEPYYNT-----FISDVVEQMQYGSWKVHFLNITYLS 800
Query: 395 EYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
E RKDGHPS +R EP P P + DC HWCLPGVPDTWNELL+
Sbjct: 799 ELRKDGHPSKYR---EPGTP-----PDAPQDCSHWCLPGVPDTWNELLY 677
>BU494295
Length = 403
Score = 77.8 bits (190), Expect = 3e-15
Identities = 41/118 (34%), Positives = 62/118 (51%), Gaps = 1/118 (0%)
Frame = +1
Query: 289 DEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPESEGN 348
DE + +E+ A E + W+ WV +DP + +VFF +MSP H+ S WN +
Sbjct: 43 DEGSTEYDEIPRPIAYERVLKTWSKWVDDNIDPNRTKVFFSSMSPLHIKSEAWNNPNGIK 222
Query: 349 CYGEKKPI-DVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIF 405
C E PI ++ + G+D N+ S+N V+ LN+T LSE RKD H S++
Sbjct: 223 CAKETTPILNMSTPLDVGTDRRLYVIANNVTHSMNVPVNFLNITSLSELRKDAHTSVY 396
>TC20057 similar to UP|O80872 (O80872) At2g30010 protein
(At2g30010/F23F1.7), partial (15%)
Length = 484
Score = 66.6 bits (161), Expect(2) = 5e-15
Identities = 30/73 (41%), Positives = 44/73 (60%)
Frame = +1
Query: 371 MSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWC 430
M V+ ++ + + ++++T LS RKD HPS++ L P +NP YSDC WC
Sbjct: 148 MKVVDIVIREMRNPAYLMDITMLSALRKDAHPSVYSG---DLSPAPRANPD-YSDCSPWC 315
Query: 431 LPGVPDTWNELLF 443
LPG+PD WNEL +
Sbjct: 316 LPGLPDAWNELFY 354
Score = 30.8 bits (68), Expect(2) = 5e-15
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 4/34 (11%)
Frame = +2
Query: 327 FFVTMSPTHLWSREWNPE----SEGNCYGEKKPI 356
FF+ +SP+H EW+ + NCYGE PI
Sbjct: 2 FFLGISPSHTHPNEWSSGVTTVTSKNCYGETAPI 103
>AI967462
Length = 464
Score = 75.1 bits (183), Expect = 2e-14
Identities = 45/157 (28%), Positives = 77/157 (48%), Gaps = 2/157 (1%)
Frame = +1
Query: 222 YNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG 281
Y +++ P L + +D ++ DS+ + A+ W D+L+FN++ WW
Sbjct: 16 YGVSIQLYRTPYLVDIVREDV------GAVLALDSI-QQANAWTDLDMLIFNSWHWWTHT 174
Query: 282 PVKLLWTDEENGA--CEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSR 339
W NG+ +++D A + WA WV VDP K +VFF +SPTH +
Sbjct: 175 GDSQGWDYIRNGSNLVKDMDRLEAFYQGLTTWARWVDLNVDPTKTKVFFQGISPTHYQGQ 354
Query: 340 EWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVEN 376
EWN + + +C G+ +P+ SY + LP +++ N
Sbjct: 355 EWN-QPKKSCSGQIEPLSGSSY---PAGLPPPTTIVN 453
>CB829368
Length = 541
Score = 74.7 bits (182), Expect = 3e-14
Identities = 45/143 (31%), Positives = 73/143 (50%), Gaps = 1/143 (0%)
Frame = +2
Query: 95 SKSSSNRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGY-QHWR 153
S SSS+ S C+++ G WVFD PLY+++ CP+ + C ++ R +L + W+
Sbjct: 89 SYSSSSSSTSISKCNLYRGNWVFDPDRTPLYDDT-CPFHRNAWNCIRNQRQNLSFINSWK 265
Query: 154 WQPHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAH 213
W P CDL R + +R K + FVGDSLN S +C+L+ ++ A
Sbjct: 266 WVPRGCDLPRIDPFRFLGLMRNKNVGFVGDSLNENFLASFLCILRVADKGARKWKKKGAW 445
Query: 214 LTIFRAEEYNATVEFLWAPLLAE 236
+ ++N TV + A LL++
Sbjct: 446 RGAY-FPKFNVTVGYHRAVLLSK 511
>BP051681
Length = 586
Score = 74.3 bits (181), Expect = 4e-14
Identities = 40/104 (38%), Positives = 59/104 (56%)
Frame = -3
Query: 339 REWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRK 398
+EWN + + +C G+ +P+ SY +G PT + V +L ++ V +L++T LS+ RK
Sbjct: 506 QEWN-QPKKSCSGQIEPLSGSSY-PAGLPPPT-TIVNKVLKNMKKPVYLLDITLLSQLRK 336
Query: 399 DGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELL 442
D HPS + +DC HWCLPG+PDTWNELL
Sbjct: 335 DAHPSAY------------GGNQGGNDCSHWCLPGLPDTWNELL 240
>TC13763 similar to UP|O80855 (O80855) At2g30900 protein, partial (13%)
Length = 538
Score = 72.0 bits (175), Expect = 2e-13
Identities = 36/97 (37%), Positives = 57/97 (58%), Gaps = 1/97 (1%)
Frame = +3
Query: 348 NCYGEKKPIDVESYWGSGSDLPTMSSVEN-ILSSLNSKVSVLNVTQLSEYRKDGHPSIFR 406
+C G+ +P+ +Y + LP +++ N +L +N+ V +L++T LS+ RKD HPS +
Sbjct: 6 SCSGQLEPLAGSTY---PAGLPQAANILNSVLKKMNNPVYLLDITLLSQLRKDAHPSTY- 173
Query: 407 KFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
S +DC HWCLPG+PDTWN+LL+
Sbjct: 174 -----------GEDHSGNDCSHWCLPGLPDTWNQLLY 251
>AV423696
Length = 427
Score = 66.6 bits (161), Expect = 8e-12
Identities = 31/98 (31%), Positives = 53/98 (53%)
Frame = +1
Query: 139 CNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQ 198
C +GR D + +WRW+P++C++ R+ + +R K + FVGDSL R Q S++C+L
Sbjct: 34 CITNGRPDSDFLYWRWKPNECNIPRFEPHTFLQLIRNKHVAFVGDSLARNQLESLLCMLS 213
Query: 199 SEIPADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAE 236
PA + + + +NAT+ W+P L +
Sbjct: 214 ---PASTLKLLHHKGARWWHFPSHNATLSSYWSPFLVQ 318
>TC11504 similar to UP|O82273 (O82273) At2g31110 protein, partial (40%)
Length = 579
Score = 63.2 bits (152), Expect = 9e-11
Identities = 30/69 (43%), Positives = 39/69 (56%)
Frame = +3
Query: 374 VENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPG 433
V +L+ ++ V L+VT LS+YRKD HP + +DC HWCLPG
Sbjct: 51 VNKVLTRMSKPVYFLDVTTLSQYRKDAHPEGYSGVMA-------------TDCSHWCLPG 191
Query: 434 VPDTWNELL 442
+PDTWNELL
Sbjct: 192 LPDTWNELL 218
>AV421063
Length = 399
Score = 60.8 bits (146), Expect = 4e-10
Identities = 28/59 (47%), Positives = 36/59 (60%)
Frame = +2
Query: 385 VSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
V +L++T LS+ R +GH S F +P DC+HWCLPGVPDTWNE+LF
Sbjct: 89 VKLLDITGLSQVRDEGHISRFSMTAKP----------GVQDCLHWCLPGVPDTWNEILF 235
>BP048416
Length = 509
Score = 56.2 bits (134), Expect = 1e-08
Identities = 39/131 (29%), Positives = 63/131 (47%), Gaps = 10/131 (7%)
Frame = -1
Query: 326 VFFVTMSPTHLWSREWNP-----ESEGNCYGEK--KPIDVESYWGSGSDLPTMSSVENIL 378
V T SP+H + +W+ +SE GEK + +D E ++ + S +
Sbjct: 503 VIMRTFSPSH-FKGDWDKGGTCAKSEPYRKGEKXLEGMDAEI---RRIEMEEVESAKGKA 336
Query: 379 SSLNSKVSVLNVTQLSEYRKDGHPSIFRK---FWEPLRPEQLSNPSSYSDCIHWCLPGVP 435
L ++ +++VT+L+ R DGHP + K F + E + SDC+HWC+PG
Sbjct: 335 KQLGFRLELMDVTELALLRPDGHPGAYMKPFPFANGVVQEHVQ-----SDCVHWCVPGPI 171
Query: 436 DTWNELLFHFL 446
DTWNE +
Sbjct: 170 DTWNEFFLEIM 138
>TC16575 homologue to GB|AAB61094.1|2194119|F20P5 F20P5.5 {Arabidopsis
thaliana;}, partial (8%)
Length = 868
Score = 55.8 bits (133), Expect = 1e-08
Identities = 38/129 (29%), Positives = 56/129 (42%), Gaps = 9/129 (6%)
Frame = +2
Query: 327 FFVTMSPTHLWSREWNPESEGNCYGEK---------KPIDVESYWGSGSDLPTMSSVENI 377
F T SP+H + WN GNC K + +++E Y L E
Sbjct: 290 FLRTFSPSHFENGIWN--EGGNCVRTKPFRSNETQLEGLNLEYYM---IQLEEFKIAEKE 454
Query: 378 LSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDT 437
+ + + Q + R DGHPS + + P + N + Y+DC+HWCLPG DT
Sbjct: 455 ARKKGLRYRLFDTMQATLLRPDGHPSKYGHW-----PNE--NVTLYNDCVHWCLPGPIDT 613
Query: 438 WNELLFHFL 446
W++ L L
Sbjct: 614 WSDFLLDLL 640
>AV765128
Length = 506
Score = 55.1 bits (131), Expect = 2e-08
Identities = 25/66 (37%), Positives = 38/66 (56%), Gaps = 3/66 (4%)
Frame = -1
Query: 384 KVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSS---YSDCIHWCLPGVPDTWNE 440
++ LNVTQL+ R +GHP + ++P +N +DC+HWCLPG DTWNE
Sbjct: 398 RLEALNVTQLALLRPNGHPGAY------IKPFPFANGVQERVQNDCVHWCLPGPIDTWNE 237
Query: 441 LLFHFL 446
+ + +
Sbjct: 236 IFLYMV 219
>BP054220
Length = 424
Score = 53.9 bits (128), Expect = 5e-08
Identities = 25/64 (39%), Positives = 38/64 (59%), Gaps = 5/64 (7%)
Frame = -3
Query: 388 LNVTQLSEYRKDGHPSIFRKFWEPLR-----PEQLSNPSSYSDCIHWCLPGVPDTWNELL 442
+++T+ +YR DGHP +R +P + P+ P DC+HWC+PG DTWNEL+
Sbjct: 422 MDITEAFQYRHDGHPGPYRSP-DPNKLTKRGPDGRPPPQ---DCLHWCMPGPVDTWNELV 255
Query: 443 FHFL 446
F +
Sbjct: 254 FEII 243
>CB827623
Length = 419
Score = 53.9 bits (128), Expect = 5e-08
Identities = 24/71 (33%), Positives = 39/71 (54%)
Frame = +2
Query: 95 SKSSSNRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRW 154
++S+S S + CD+F+G+WV N P Y C + + C K+GR D + WRW
Sbjct: 194 NESASLPSTSVKKCDIFTGEWV-PNPKAPYYTNKTCWAIHEHQNCMKYGRPDSEFMKWRW 370
Query: 155 QPHDCDLKRWN 165
+P+ +L +N
Sbjct: 371 KPNGXELPIFN 403
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.134 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,396,824
Number of Sequences: 28460
Number of extensions: 175469
Number of successful extensions: 1093
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 1070
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1077
length of query: 446
length of database: 4,897,600
effective HSP length: 93
effective length of query: 353
effective length of database: 2,250,820
effective search space: 794539460
effective search space used: 794539460
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC143338.3


