

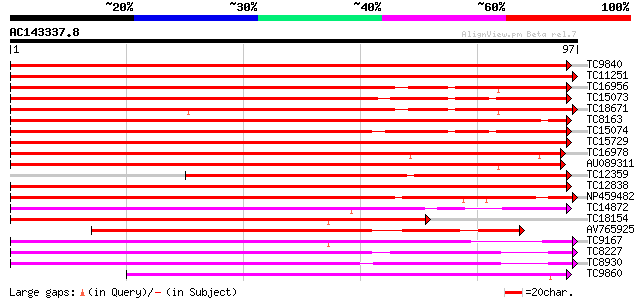
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.8 - phase: 0
(97 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9840 UP|R11D_LOTJA (Q40194) Ras-related protein Rab11D, complete 176 5e-46
TC11251 UP|R11E_LOTJA (Q40195) Ras-related protein Rab11E, complete 157 2e-40
TC16956 homologue to UP|Q9FJH0 (Q9FJH0) GTP-binding protein, ras... 119 9e-29
TC15073 UP|Q40199 (Q40199) RAB11I (Fragment), complete 110 4e-26
TC18671 UP|Q40198 (Q40198) RAB11H, complete 107 4e-25
TC8163 UP|R11C_LOTJA (Q40193) Ras-related protein Rab11C, complete 105 1e-24
TC15074 similar to UP|R11B_TOBAC (Q40521) Ras-related protein Ra... 103 5e-24
TC15729 homologue to UP|RB1C_ARATH (O04486) Ras-related protein ... 92 1e-20
TC16978 UP|R11A_LOTJA (Q40191) Ras-related protein Rab11A, complete 92 2e-20
AU089311 89 1e-19
TC12359 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, p... 88 3e-19
TC12838 UP|Q40196 (Q40196) RAB11F, complete 79 1e-16
NP459482 RAB11J [Lotus japonicus] 70 5e-14
TC14872 UP|Q40208 (Q40208) RAB2A, complete 62 2e-11
TC18154 UP|Q40192 (Q40192) RAB11B, complete 59 2e-10
AV765925 57 5e-10
TC9167 UP|Q40202 (Q40202) RAB1B (Fragment), complete 55 3e-09
TC8227 UP|Q40203 (Q40203) RAB1C, complete 54 6e-09
TC8930 UP|Q40204 (Q40204) RAB1D, complete 51 3e-08
TC9860 UP|Q40197 (Q40197) RAB11G, complete 51 3e-08
>TC9840 UP|R11D_LOTJA (Q40194) Ras-related protein Rab11D, complete
Length = 1302
Score = 176 bits (447), Expect = 5e-46
Identities = 89/96 (92%), Positives = 93/96 (96%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVL+QIYRIVSK+
Sbjct: 611 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLTQIYRIVSKR 790
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
AVEAG+ GSSS +PS GQTINVKEDSSVLKRFGCCS
Sbjct: 791 AVEAGDSGSSSGLPSKGQTINVKEDSSVLKRFGCCS 898
>TC11251 UP|R11E_LOTJA (Q40195) Ras-related protein Rab11E, complete
Length = 950
Score = 157 bits (398), Expect = 2e-40
Identities = 80/97 (82%), Positives = 88/97 (90%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNKSDLRHLV V TEDGK+FAE+ESLYFMETSALEATNVENAF+EVL+QIYRIVSK+
Sbjct: 425 MLIGNKSDLRHLVTVSTEDGKAFAEKESLYFMETSALEATNVENAFSEVLTQIYRIVSKR 604
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
AVEAG+ S+S VPS GQTINV EDSSVL R+ CCSN
Sbjct: 605 AVEAGDRPSTSVVPSQGQTINVNEDSSVLNRYRCCSN 715
>TC16956 homologue to UP|Q9FJH0 (Q9FJH0) GTP-binding protein, ras-like
(At5g60860), partial (44%)
Length = 584
Score = 119 bits (298), Expect = 9e-29
Identities = 62/98 (63%), Positives = 79/98 (80%), Gaps = 2/98 (2%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL AV T+D K FAERE+ +FMETSALE+ NV+NAFTEVL+QIYR+VSKK
Sbjct: 6 MLVGNKADLRHLRAVSTDDAKGFAERENTFFMETSALESLNVDNAFTEVLTQIYRVVSKK 185
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCS 96
+E G+ +++P GQTINV K+D S +K+ GCCS
Sbjct: 186 TLEIGD--DPAALPK-GQTINVGSKDDVSAVKKVGCCS 290
>TC15073 UP|Q40199 (Q40199) RAB11I (Fragment), complete
Length = 733
Score = 110 bits (275), Expect = 4e-26
Identities = 58/96 (60%), Positives = 75/96 (77%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL AVPTE+ +FAERE+ YFMETSALE+ NV+NAF EVLS+IY +VS+K
Sbjct: 211 MLVGNKADLRHLRAVPTEEATAFAERENTYFMETSALESLNVDNAFIEVLSEIYNVVSRK 390
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
+E +G ++P GQTIN+ D S +K+ GCCS
Sbjct: 391 TLE--KGNDPGALPQ-GQTINL-GDVSAVKKPGCCS 486
>TC18671 UP|Q40198 (Q40198) RAB11H, complete
Length = 1181
Score = 107 bits (266), Expect = 4e-25
Identities = 60/100 (60%), Positives = 76/100 (76%), Gaps = 3/100 (3%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESL-YFMETSALEATNVENAFTEVLSQIYRIVSK 59
ML+GNK+DLRHL AV TED F E +FMETSALE+ NVENAFTEVL+QIYR+VSK
Sbjct: 567 MLVGNKADLRHLRAVSTEDSSGFLLNERTHFFMETSALESLNVENAFTEVLTQIYRVVSK 746
Query: 60 KAVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCSN 97
KA+E G+ +++P GQTINV ++D S +K+ GCCS+
Sbjct: 747 KALEIGD--DPTALPK-GQTINVGSRDDVSPVKKSGCCSS 857
>TC8163 UP|R11C_LOTJA (Q40193) Ras-related protein Rab11C, complete
Length = 1193
Score = 105 bits (263), Expect = 1e-24
Identities = 55/96 (57%), Positives = 69/96 (71%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
M+ GNKSDL HL AV +DG + AE+E L F+ETSALEATN+E AF +L++IY IVSKK
Sbjct: 597 MMAGNKSDLNHLRAVSEDDGGALAEKEGLSFLETSALEATNIEKAFQTILTEIYHIVSKK 776
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
A+ A E + +SVP G TINV + S K+ GCCS
Sbjct: 777 ALAAQEATAGASVPGQGTTINVADTSGNTKK-GCCS 881
>TC15074 similar to UP|R11B_TOBAC (Q40521) Ras-related protein Rab11B,
partial (89%)
Length = 973
Score = 103 bits (257), Expect = 5e-24
Identities = 56/96 (58%), Positives = 71/96 (73%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL AVPTE+ +FAERE+ YFMETSALE+ NV+NAF VLS+IY +VS+K
Sbjct: 525 MLVGNKADLRHLRAVPTEEATAFAERENTYFMETSALESLNVDNAFIXVLSEIYNVVSRK 704
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
+ +G ++P GQTIN+ D S K GCCS
Sbjct: 705 TL--XKGNDPGALPQ-GQTINL-GDVSXXKXPGCCS 800
>TC15729 homologue to UP|RB1C_ARATH (O04486) Ras-related protein Rab11C,
partial (85%)
Length = 860
Score = 92.4 bits (228), Expect = 1e-20
Identities = 47/96 (48%), Positives = 64/96 (65%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK+DL+HL V TED +S+AE+E L F+ETSALEATNV+ AF +L+QIYRI+SKK
Sbjct: 443 MLIGNKTDLKHLRGVATEDAQSYAEKEGLSFIETSALEATNVDKAFQTILAQIYRIISKK 622
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
++ + S+ G TI ++ CC+
Sbjct: 623 SLSSSSTDPSAPNIKQGNTITIQGGLQPNTSKSCCT 730
>TC16978 UP|R11A_LOTJA (Q40191) Ras-related protein Rab11A, complete
Length = 984
Score = 91.7 bits (226), Expect = 2e-20
Identities = 52/98 (53%), Positives = 68/98 (69%), Gaps = 3/98 (3%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+LIGNK DL + VPTED K FAE+E L+F+ETSALEATNVE+AFT VL++IY IV+KK
Sbjct: 437 ILIGNKCDLVNQRDVPTEDAKEFAEKEGLFFLETSALEATNVESAFTTVLTEIYNIVNKK 616
Query: 61 AVEAGEG-GSSSSVPSVGQTINVKEDSSVL--KRFGCC 95
++ A E G+ +S GQ I + + + KR CC
Sbjct: 617 SLAADESQGNGNSASLSGQKIIIPGPAQEIPAKRNMCC 730
>AU089311
Length = 344
Score = 89.4 bits (220), Expect = 1e-19
Identities = 50/97 (51%), Positives = 63/97 (64%), Gaps = 2/97 (2%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK DL L AVPTED + FA+RE+L+FMETSALE+TNVE F +L++IYRI +KK
Sbjct: 35 MLIGNKCDLGSLRAVPTEDAEEFAQRENLFFMETSALESTNVETCFLTILTEIYRINAKK 214
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCC 95
+ G+ S G I V +E K+ GCC
Sbjct: 215 TLTVGDDPDGGSGLLKGSRIIVPNQEMDGGGKKGGCC 325
>TC12359 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, partial
(30%)
Length = 360
Score = 87.8 bits (216), Expect = 3e-19
Identities = 44/66 (66%), Positives = 54/66 (81%)
Frame = +1
Query: 31 FMETSALEATNVENAFTEVLSQIYRIVSKKAVEAGEGGSSSSVPSVGQTINVKEDSSVLK 90
F+ETSALEA+NVENAF EVL+QIY +VSKKAVE E G ++SVP+ G+ I++K D S LK
Sbjct: 1 FIETSALEASNVENAFAEVLTQIYNVVSKKAVEGAENG-TASVPAKGEKIDLKNDVSALK 177
Query: 91 RFGCCS 96
R GCCS
Sbjct: 178RVGCCS 195
>TC12838 UP|Q40196 (Q40196) RAB11F, complete
Length = 1099
Score = 79.0 bits (193), Expect = 1e-16
Identities = 40/96 (41%), Positives = 62/96 (63%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+ NKSDL V E+G+ FAE E L FMETSAL+ NVE AF E++++I+ I+S+K
Sbjct: 473 VLVANKSDLVQSREVEKEEGEEFAEIEGLCFMETSALQNLNVEEAFLEMITKIHDIISQK 652
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
++E G ++ +G+ I V ++ + K+ CCS
Sbjct: 653 SLETKMNGPTTLSVPIGKEIQVADEVTATKQANCCS 760
>NP459482 RAB11J [Lotus japonicus]
Length = 672
Score = 70.5 bits (171), Expect = 5e-14
Identities = 42/104 (40%), Positives = 66/104 (63%), Gaps = 7/104 (6%)
Frame = +1
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNKSDL+ V T +GK+ AE + L+FMETSAL+++NV AF V+ +IY I+S+K
Sbjct: 367 ILVGNKSDLKDAREVTTAEGKALAEAQGLFFMETSALDSSNVAAAFQTVVKEIYNILSRK 546
Query: 61 AVEAGEGGSSSSVPSV--GQTI-----NVKEDSSVLKRFGCCSN 97
+ + E + VP + G+++ N++ D K GCCS+
Sbjct: 547 VMMSQE-LTKHEVPRIENGKSVVIQGENLEADGQSKK--GCCSS 669
>TC14872 UP|Q40208 (Q40208) RAB2A, complete
Length = 1125
Score = 61.6 bits (148), Expect = 2e-11
Identities = 41/105 (39%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIV--- 57
MLIGNK DL H AV TE+G+ FA+ L FME SA A NVE AF + + IY+ +
Sbjct: 549 MLIGNKCDLAHRRAVSTEEGEQFAKEHGLIFMEASAKTAQNVEEAFIKTAATIYKKIQDG 728
Query: 58 ------SKKAVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
++ G GG PS G+ D GCCS
Sbjct: 729 VFDVSNESYGIKVGYGGIPG--PSGGR------DGPSASAGGCCS 839
>TC18154 UP|Q40192 (Q40192) RAB11B, complete
Length = 1183
Score = 58.5 bits (140), Expect = 2e-10
Identities = 33/73 (45%), Positives = 46/73 (62%), Gaps = 1/73 (1%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQI-YRIVSK 59
MLIGNK DL V +ED FAE + L+F ETSAL NVE+AF ++L +I R+VSK
Sbjct: 695 MLIGNKGDLVDQRVVLSEDAVEFAEEQGLFFSETSALTGENVESAFLKLLQEINTRVVSK 874
Query: 60 KAVEAGEGGSSSS 72
+++ G ++
Sbjct: 875 RSLSDCNHGKKTN 913
>AV765925
Length = 374
Score = 57.0 bits (136), Expect = 5e-10
Identities = 34/74 (45%), Positives = 49/74 (65%)
Frame = -3
Query: 15 VPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKKAVEAGEGGSSSSVP 74
V ED +S+AE+E L F++TSALEATNV AF +L++I RI SKK++ SSS+ P
Sbjct: 369 VAPEDAQSYAEKEGLSFIKTSALEATNVAKAFQPILARINRIPSKKSL-----SSSSTDP 205
Query: 75 SVGQTINVKEDSSV 88
S N+K+ + +
Sbjct: 204 SAP---NIKQGNPI 172
>TC9167 UP|Q40202 (Q40202) RAB1B (Fragment), complete
Length = 951
Score = 54.7 bits (130), Expect = 3e-09
Identities = 39/98 (39%), Positives = 52/98 (52%), Gaps = 1/98 (1%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQI-YRIVSK 59
+L+GNKSDL AV E K+FA+ + FMETSA +ATNVE AF + + I R+ S+
Sbjct: 440 LLVGNKSDLTANRAVSYETAKAFADEIGIPFMETSAKDATNVEQAFMAMAASIKNRMASQ 619
Query: 60 KAVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
A A + S VGQ+ GCCS+
Sbjct: 620 PANNARPPTVNISGKPVGQS------------SGCCSS 697
>TC8227 UP|Q40203 (Q40203) RAB1C, complete
Length = 1083
Score = 53.5 bits (127), Expect = 6e-09
Identities = 35/97 (36%), Positives = 49/97 (50%)
Frame = +2
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNK DL V E K+FA+ + FMETSA ATNVE AF + ++I ++ +
Sbjct: 476 LLVGNKCDLTANKVVSYETAKAFADEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQ 655
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
V +V GQ +N K GCCS+
Sbjct: 656 PV---NNARPPTVQIRGQPVNQKS--------GCCSS 733
>TC8930 UP|Q40204 (Q40204) RAB1D, complete
Length = 943
Score = 51.2 bits (121), Expect = 3e-08
Identities = 34/97 (35%), Positives = 49/97 (50%)
Frame = +3
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+L+GNK DL AV + K FA+ + FMETSA +ATNVE AF + + I ++ +
Sbjct: 438 LLVGNKCDLTANRAVSYDTAKEFADPIGIPFMETSAKDATNVEQAFMAMSASIKNRMASQ 617
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
+ G +V GQ + K GCCS+
Sbjct: 618 --PSANNGRPPTVQIRGQPVGQKS--------GCCSS 698
>TC9860 UP|Q40197 (Q40197) RAB11G, complete
Length = 1104
Score = 51.2 bits (121), Expect = 3e-08
Identities = 30/77 (38%), Positives = 42/77 (53%), Gaps = 1/77 (1%)
Frame = +2
Query: 21 KSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKKAVEAGEGGSSSSVPSVGQTI 80
K+ + + L+FMETSAL++TNV AF V+ +IY VS+K + + + SV V
Sbjct: 521 KALQKAQGLFFMETSALDSTNVRTAFEMVIREIYNNVSRKVLNSDTYKAELSVDRVSLVN 700
Query: 81 NVKEDSSVLKR-FGCCS 96
N S K F CCS
Sbjct: 701 NGAATSKQSKSYFSCCS 751
Score = 30.8 bits (68), Expect = 0.040
Identities = 15/28 (53%), Positives = 18/28 (63%)
Frame = +1
Query: 4 GNKSDLRHLVAVPTEDGKSFAERESLYF 31
GNK DL ++ AV E+GKS AE L F
Sbjct: 469 GNKCDLENIRAVSIEEGKSLAEGARLVF 552
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.127 0.336
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,257,985
Number of Sequences: 28460
Number of extensions: 9653
Number of successful extensions: 75
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 71
length of query: 97
length of database: 4,897,600
effective HSP length: 73
effective length of query: 24
effective length of database: 2,820,020
effective search space: 67680480
effective search space used: 67680480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 47 (22.7 bits)
Medicago: description of AC143337.8


