

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.12 - phase: 0
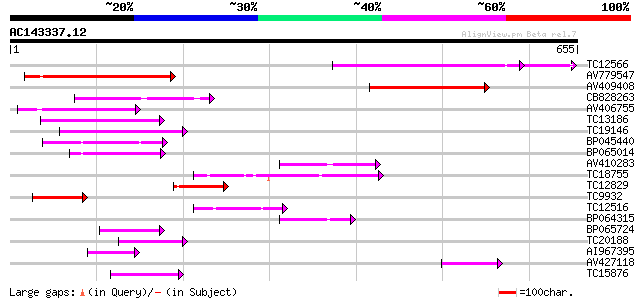
(655 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (... 163 2e-45
AV779547 151 4e-37
AV409408 129 2e-30
CB828263 111 3e-25
AV406755 110 5e-25
TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidat... 105 3e-23
TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-contain... 102 1e-22
BP045440 89 3e-18
BP065014 79 2e-15
AV410283 70 8e-13
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 69 2e-12
TC12829 67 1e-11
TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like... 64 8e-11
TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like pr... 62 2e-10
BP064315 61 6e-10
BP065724 59 3e-09
TC20188 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-... 52 4e-07
AI967395 51 5e-07
AV427118 49 2e-06
TC15876 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-... 47 7e-06
>TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (28%)
Length = 918
Score = 163 bits (412), Expect(2) = 2e-45
Identities = 85/222 (38%), Positives = 132/222 (59%)
Frame = +1
Query: 373 DVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRD 432
++YEV ++ D+ +LF AFK DP + L+ ++L YA+G+PLA++V+G FL R+
Sbjct: 7 EIYEVKEMSFEDSLQLFSLNAFKRNDPLETYIDLSKKLLSYAKGIPLALKVLGLFLYGRE 186
Query: 433 ANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRILDAC 492
W L +L PD +++VL++S++GL + ++IFL IACF+ G E+ V + LD+C
Sbjct: 187 RKAWESQLVKLEKLPDPEIINVLKLSYDGLDDDQKDIFLDIACFYLGHLEESVAQTLDSC 366
Query: 493 GLHPHIGIQSLIERSFITIRNNEILMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYS 552
G IG++ L +R I+I I+MH++++ +GK+IVRQQ P PG SRLW D Y
Sbjct: 367 GFFADIGMEVLKDRCLISILEGRIVMHDLIEVMGKEIVRQQCPNDPGKRSRLWNDDQTYD 546
Query: 553 VMMTETGTNNINAIILDQKEHISEYPQLRAEALSIMRGLKIL 594
V+ GT+ I I L + L AE M L++L
Sbjct: 547 VLGKNKGTDAIQCIFLYTCP--IKIVNLHAETFKYMTNLRML 666
Score = 37.4 bits (85), Expect(2) = 2e-45
Identities = 23/66 (34%), Positives = 35/66 (52%), Gaps = 2/66 (3%)
Frame = +2
Query: 591 LKILILLFHKN--FSGSLTFLSNSLQYLLWYGYPFASLPLNFEPFCLVELNMPYSSIQRL 648
+ ++ + H N SL L N LQ+L W G+P PL+F P LV+L+M S ++
Sbjct: 674 ISLVSVPLHSNVILPASLGSLPNHLQFLRWDGFPLRYFPLDFCPQNLVKLDMRDSHLEHF 853
Query: 649 WDGHKV 654
G K+
Sbjct: 854 --GRKI 865
>AV779547
Length = 556
Score = 151 bits (381), Expect = 4e-37
Identities = 81/174 (46%), Positives = 107/174 (60%)
Frame = +1
Query: 18 IHLGQGDIGSSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLE 77
+H G G+ +S+ ++ +SFRG DTR+ F DHL L KG FKDD L
Sbjct: 43 LHFGNGNPKQLFNSDDME---I*CVVSFRGEDTRNNFTDHLLGALYGKGFVTFKDDTMLR 213
Query: 78 KGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADP 137
KG+++S +L+QAI+ S+I IVVFSK YA ST CL+E+A IA+ +QTV P+ D P
Sbjct: 214 KGKNISTELIQAIEGSQILIVVFSKYYASSTWCLQELAKIADGIVGKRQTVLPVVCDVTP 393
Query: 138 SHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAKLVGWDVRNKPEFRE 191
S VRKQSG Y AF+ + +FK D V RW A+ +A L GWDV NKPE +
Sbjct: 394 SEVRKQSGNYGEAFLKHEERFKEDLEMVQRWRKALAEVADLSGWDVTNKPEHEQ 555
>AV409408
Length = 430
Score = 129 bits (323), Expect = 2e-30
Identities = 61/139 (43%), Positives = 92/139 (65%)
Frame = +2
Query: 416 GLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIAC 475
GLPLA+ V+GSFLC R + W DAL +++ P +++++ L++S++ L E + +FL IAC
Sbjct: 5 GLPLALEVLGSFLCGRSLSDWEDALIKIKKVPHDDILNKLRISYDMLEDEHKTLFLDIAC 184
Query: 476 FFKGEKEDYVKRILDACGLHPHIGIQSLIERSFITIRNNEILMHEMLQELGKKIVRQQFP 535
FFKG + V ++L +CGLHP +GI LIE+S +T I MH++L+E+GK V + P
Sbjct: 185 FFKGWYKHKVIQLLGSCGLHPTVGINVLIEKSLLTFDGRVIGMHDLLEEMGKTEVFLESP 364
Query: 536 FQPGSWSRLWLYDDFYSVM 554
PG SRLW +D V+
Sbjct: 365 NDPGRRSRLWSLEDIDQVL 421
>CB828263
Length = 570
Score = 111 bits (278), Expect = 3e-25
Identities = 61/163 (37%), Positives = 93/163 (56%), Gaps = 2/163 (1%)
Frame = +2
Query: 76 LEKGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDA 135
L++G+ +S LL+AI+ +++S++VFSKNY S CL+E+ I E Q V P+FYD
Sbjct: 8 LQRGDEISSSLLRAIEEAKLSVIVFSKNYGNSKWCLDELVKILECKRTRGQIVLPVFYDI 187
Query: 136 DPSHVRKQSGVYQNAFVLLQNKFKH-DPNKVMRWVGAMESLAKLVGWDVR-NKPEFREIK 193
DPSHVR Q+G Y AFV KH +KV +W A+ A L GWD N+ E I+
Sbjct: 188 DPSHVRNQTGTYAEAFV------KHGQVDKVQKWREALREAANLSGWDCSVNRMESEVIE 349
Query: 194 NIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYE 236
I ++V+ + ++G D+ ++ + E L +L + YE
Sbjct: 350 KIAKDVLEKLNRVYVGDLDE------KIAKFEQLAQLQREFYE 460
>AV406755
Length = 425
Score = 110 bits (276), Expect = 5e-25
Identities = 60/143 (41%), Positives = 87/143 (59%), Gaps = 1/143 (0%)
Frame = +3
Query: 10 SRLTSSDLI-HLGQGDIGSSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIF 68
++L DL+ +G+G I S + YDVF+SFRG ++R +F HL+ L GI
Sbjct: 6 NKLQMHDLLKEMGRGIIVKKPKSK----WSYDVFLSFRGEESRRSFTSHLYTALKNAGIK 173
Query: 69 AFKDDKRLEKGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTV 128
F D++ L++GE +S LL+AI+ SRI+I++FS NY S CL+E+ I E + Q V
Sbjct: 174 VFMDNE-LQRGEDISSSLLKAIEDSRIAIIIFSTNYTGSKWCLDELEKIIECQRTIGQEV 350
Query: 129 FPIFYDADPSHVRKQSGVYQNAF 151
P+FY+ DPS +RKQ G AF
Sbjct: 351 MPVFYNVDPSDIRKQRGTVGEAF 419
Score = 32.7 bits (73), Expect = 0.19
Identities = 26/103 (25%), Positives = 46/103 (44%), Gaps = 10/103 (9%)
Frame = +3
Query: 513 NNEILMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTE----------TGTNN 562
NN++ MH++L+E+G+ I+ + +P S W YD F S E T N
Sbjct: 3 NNKLQMHDLLKEMGRGIIVK----KPKS---KWSYDVFLSFRGEESRRSFTSHLYTALKN 161
Query: 563 INAIILDQKEHISEYPQLRAEALSIMRGLKILILLFHKNFSGS 605
+ E + + + L + +I I++F N++GS
Sbjct: 162 AGIKVFMDNE-LQRGEDISSSLLKAIEDSRIAIIIFSTNYTGS 287
>TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (6%)
Length = 586
Score = 105 bits (261), Expect = 3e-23
Identities = 58/143 (40%), Positives = 83/143 (57%)
Frame = +1
Query: 36 SYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRI 95
S+ YDVFIS +DTR F L+ L+ KGI F DD + +S +AI+SSRI
Sbjct: 94 SFIYDVFISLITSDTRFGFSGDLYTALSDKGILTFIDDGLPRPDDIMSTVNSKAIESSRI 273
Query: 96 SIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQ 155
+IVV S+NYA S+ CL+E+ I + E ++P+FY +PS VR Q G + AF +
Sbjct: 274 AIVVISENYASSSFCLDELMYILKRREEKGLLIWPVFYQVEPSDVRYQKGGFGKAFASHE 453
Query: 156 NKFKHDPNKVMRWVGAMESLAKL 178
+F D KV +W A++ +A L
Sbjct: 454 ERFGSDSAKVRKWRNALKQVADL 522
>TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing protein
TSDC, partial (12%)
Length = 701
Score = 102 bits (255), Expect = 1e-22
Identities = 56/149 (37%), Positives = 86/149 (57%), Gaps = 1/149 (0%)
Frame = +1
Query: 58 LHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATI 117
L+ L +G F DD+ L G ++ LL A + SR+SI+VFS+NY S+ CL+E+ I
Sbjct: 1 LYHALRREGFKTFMDDEGLVGGNEIAKSLLDASEKSRLSIIVFSENYGYSSWCLDELVKI 180
Query: 118 AEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAK 177
E KQ V+PIFY + S V Q Y++A + + + D ++V +W A+ ++AK
Sbjct: 181 VECMETKKQLVWPIFYKVEKSDVTSQENSYEHAMIGHEKNYGKDSDEVKKWRSALSAMAK 360
Query: 178 LVGWDVR-NKPEFREIKNIVQEVINTMGH 205
L G +V+ NK E+ IK IV++ I H
Sbjct: 361 LEGHNVKENKYEYEFIKKIVEKAIAIENH 447
>BP045440
Length = 535
Score = 88.6 bits (218), Expect = 3e-18
Identities = 54/147 (36%), Positives = 77/147 (51%), Gaps = 2/147 (1%)
Frame = -3
Query: 38 RYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISI 97
RY +FISFRG DTR F L+ L +G FKDD+ LE G + +LL I+ SR +I
Sbjct: 530 RYQIFISFRGMDTRDDFTKPLYQGLCREGFVTFKDDESLEGGSPIE-KLLDDIEESRFAI 354
Query: 98 VVFSKNYAESTLCLEEMATIAEYHTE--LKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQ 155
++ SKNYAES CL+E++ I E + Q V P + P+ VR Y +A +
Sbjct: 353 IILSKNYAESEWCLKELSKILECKDQEGKNQLVLPSSTEVTPTEVRHVINRYADA-MAKH 177
Query: 156 NKFKHDPNKVMRWVGAMESLAKLVGWD 182
K D + W A+ + L G++
Sbjct: 176 EKNGIDSKTIQAWKKALFDVCTLTGFE 96
>BP065014
Length = 498
Score = 79.3 bits (194), Expect = 2e-15
Identities = 42/111 (37%), Positives = 62/111 (55%)
Frame = -2
Query: 70 FKDDKRLEKGESLSPQLLQAIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVF 129
FKDD LE G + +LL AI+ SR++IVV S+N+A+S CL+E+ I + + K V
Sbjct: 497 FKDDLSLEDGAPIE-ELLGAIEESRVAIVVLSENFADSKWCLKELVKILDCRKKKKPLVL 321
Query: 130 PIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAKLVG 180
PIFY +P+++R G Y A + K D V W A+ ++ L G
Sbjct: 320 PIFYKVEPTNIRHLKGRYGEAMAEHEKKLGKDSQTVREWTQALSDISYLKG 168
>AV410283
Length = 426
Score = 70.5 bits (171), Expect = 8e-13
Identities = 43/117 (36%), Positives = 65/117 (54%)
Frame = +1
Query: 312 IRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVS 371
I++ L K L++LD+ D ++Q++ L N E KGSR++ITTR+ +L S
Sbjct: 70 IKRVLQGNKVLLILDDVDEIQQLDFLMGNREWFHKGSRVVITTRNTQVLPESYVDM---- 237
Query: 372 YDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFL 428
YEV L + A LF A + + P G L+ +++K GLPLA+ V+GSFL
Sbjct: 238 --FYEVRELELSAALALFCHHAMRRKKPAEGFSNLSKQIVKKTGGLPLALEVIGSFL 402
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 68.9 bits (167), Expect = 2e-12
Identities = 68/229 (29%), Positives = 105/229 (45%), Gaps = 9/229 (3%)
Frame = +3
Query: 213 DLIGIQPRVEELESLLKLDSKDYEFR-AIGIWGMAGIRKTTLASVLYDR--VSYQFDASC 269
DL+GI R ++L L K R I + GM G+ KTTL +YD V F A
Sbjct: 192 DLVGIDRRKKKLMGCL---IKPCPVRKVISVTGMGGMGKTTLVKQVYDDPVVIKHFRACA 362
Query: 270 FIENVSKIYKDGGATAVQKQILRQTIDE------KNLETYSPSEISGIIRKRLCNKKFLV 323
+I VS+ + G + + + RQ E LE + II+ L +++LV
Sbjct: 363 WI-TVSQSCEIG---ELLRDLARQLFSEIRRPVPLGLENMRCDRLKMIIKDLLQRRRYLV 530
Query: 324 VLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSN 383
V D+ + + E + GSRI+ITTR L+ + T VY + L +
Sbjct: 531 VFDDVWHVREWEAVKYALPDNNCGSRIMITTRRS---DLAFTSSTESKGKVYNLQPLKED 701
Query: 384 DARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRD 432
+A +LF RK F + S + + +L+ +GLPLA+ + L T+D
Sbjct: 702 EAWELFCRKTFHGDSCPSHLIGICTYILRKCEGLPLAIVAISGVLATKD 848
>TC12829
Length = 448
Score = 66.6 bits (161), Expect = 1e-11
Identities = 34/63 (53%), Positives = 45/63 (70%)
Frame = +3
Query: 190 REIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIR 249
R+ KN VQ I T+GHKF F++ L+GIQP +EE+ +LLKL +D +FR +GIW GI
Sbjct: 39 RD*KN-VQARIKTLGHKFSRFSNGLVGIQPHIEEI*NLLKLS*EDEDFRVLGIWRTGGIG 215
Query: 250 KTT 252
KTT
Sbjct: 216 KTT 224
>TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like, partial
(5%)
Length = 689
Score = 63.9 bits (154), Expect = 8e-11
Identities = 31/64 (48%), Positives = 42/64 (65%)
Frame = -2
Query: 27 SSSDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQL 86
S S S+S + YDVFI+FRG DTR +FV HL+ L+ G+ F DD+ LE+G L P+L
Sbjct: 238 SMSSSSSNHQWIYDVFINFRGKDTRHSFVSHLYTALSNAGVNVFLDDENLERGLELEPEL 59
Query: 87 LQAI 90
+ I
Sbjct: 58 QKLI 47
>TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like protein
(Fragment), partial (36%)
Length = 859
Score = 62.4 bits (150), Expect = 2e-10
Identities = 35/108 (32%), Positives = 64/108 (58%)
Frame = +3
Query: 213 DLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIE 272
+LIG+ + LESLL +SKD R IGIWGM GI KTT+A +++R Q++ CF+
Sbjct: 543 ELIGVDKSIAHLESLLCQESKDV--RVIGIWGMGGIGKTTIAEEIFNRRHSQYEGICFLS 716
Query: 273 NVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKK 320
V G ++++++ T+ ++++ +P+ ++ I++R+ K
Sbjct: 717 KVKDEL*KHGMQSLREKLF-STLLAQDVKIDTPNGMADDIKRRIGRMK 857
>BP064315
Length = 350
Score = 60.8 bits (146), Expect = 6e-10
Identities = 36/88 (40%), Positives = 52/88 (58%)
Frame = +1
Query: 312 IRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVS 371
I++RL NK+ L+VLD+ D ++Q++ LA + G GSRIIITTRD +L D V
Sbjct: 55 IKRRLSNKRVLLVLDDVDKIKQLQALAGGGDWFGSGSRIIITTRDTTMLDKHKMDD--VI 228
Query: 372 YDVYEVPLLNSNDARKLFYRKAFKSEDP 399
Y++ LN +D+ +LF AF P
Sbjct: 229 MKKYKMEELNDHDSLELFCWHAFNMSTP 312
>BP065724
Length = 495
Score = 58.5 bits (140), Expect = 3e-09
Identities = 27/75 (36%), Positives = 42/75 (56%)
Frame = -1
Query: 104 YAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPN 163
YA+S+ CLEE+ I E + Q V+PIFY +PS VR Q Y+ A + + ++ D
Sbjct: 495 YADSSWCLEEVVKILECKQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKQETRYGKDSE 316
Query: 164 KVMRWVGAMESLAKL 178
KV +W ++ + L
Sbjct: 315 KVKKWRSSLSQVCDL 271
>TC20188 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (11%)
Length = 494
Score = 51.6 bits (122), Expect = 4e-07
Identities = 27/81 (33%), Positives = 44/81 (53%), Gaps = 1/81 (1%)
Frame = +1
Query: 126 QTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLAKLVGWDV-R 184
Q V+PIFY+ S V Q+ Y +A + +N F D KV +W A+ +A L G +
Sbjct: 4 QLVWPIFYEISXSDVSNQTKSYGHAMLAHENSFGKDSEKVQKWKSALSEMAFLQGEHITE 183
Query: 185 NKPEFREIKNIVQEVINTMGH 205
N+ E++ I+ IV++ + H
Sbjct: 184 NEYEYKLIQKIVEKALAVQNH 246
>AI967395
Length = 205
Score = 51.2 bits (121), Expect = 5e-07
Identities = 25/61 (40%), Positives = 35/61 (56%)
Frame = +3
Query: 90 IQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQN 149
IQ SR+ IVVFS++YA S CL E+ I + Q V+P++Y + +R Q G Y
Sbjct: 3 IQESRLIIVVFSEHYAVSASCLNELVNIVDVMIMNNQLVWPVYYGVEAPEIRHQRGRYGE 182
Query: 150 A 150
A
Sbjct: 183 A 185
>AV427118
Length = 317
Score = 49.3 bits (116), Expect = 2e-06
Identities = 26/71 (36%), Positives = 42/71 (58%), Gaps = 1/71 (1%)
Frame = +3
Query: 500 IQSLIERSFITIRNN-EILMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTET 558
I +L+ + I I ++ E+ +HE ++ +GK+IVR++ P SRLW+ D V+ T
Sbjct: 54 ISALVSKFLINISSSGELTLHEWMRNMGKEIVRRKSSRMPCVSSRLWILQDIRQVLEDCT 233
Query: 559 GTNNINAIILD 569
GT+ I I LD
Sbjct: 234 GTHKIRTICLD 266
>TC15876 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (13%)
Length = 558
Score = 47.4 bits (111), Expect = 7e-06
Identities = 24/87 (27%), Positives = 44/87 (49%), Gaps = 2/87 (2%)
Frame = +1
Query: 117 IAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRWVGAMESLA 176
I E + Q V+PIFY +PS VR Q Y+ A + + ++ D KV +W ++ +
Sbjct: 16 ILECXQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKPETRYGKDSEKVKKWRSSLSQVC 195
Query: 177 KLVGWDVRNKPEFRE--IKNIVQEVIN 201
L + + + I +I+++V+N
Sbjct: 196 DLKAFHYKENSGYERAFIPDIIKDVMN 276
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,933,711
Number of Sequences: 28460
Number of extensions: 142473
Number of successful extensions: 716
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 706
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 710
length of query: 655
length of database: 4,897,600
effective HSP length: 96
effective length of query: 559
effective length of database: 2,165,440
effective search space: 1210480960
effective search space used: 1210480960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC143337.12


