

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142506.2 + phase: 1 /partial
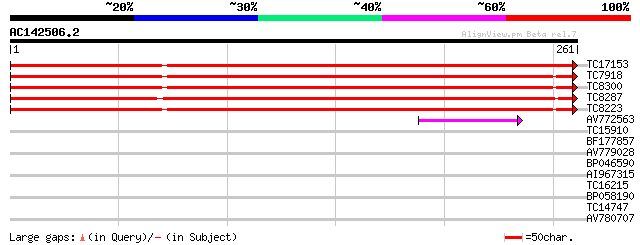
(261 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17153 UP|AAQ87023 (AAQ87023) VDAC3.1, complete 421 e-119
TC7918 UP|AAQ87019 (AAQ87019) VDAC1.1, complete 226 3e-60
TC8300 UP|AAQ87020 (AAQ87020) VDAC1.2, complete 217 2e-57
TC8287 UP|AAQ87022 (AAQ87022) VDAC2.1, complete 216 4e-57
TC8223 UP|AAQ87021 (AAQ87021) VDAC1.3, complete 213 3e-56
AV772563 40 4e-04
TC15910 similar to UP|Q9AR64 (Q9AR64) Urease accessory protein G... 29 0.93
BF177857 27 2.7
AV779028 27 2.7
BP046590 27 3.5
AI967315 27 4.6
TC16215 weakly similar to UP|Q9FI12 (Q9FI12) Serine proteinase, ... 27 4.6
BP058190 27 4.6
TC14747 26 6.1
AV780707 26 7.9
>TC17153 UP|AAQ87023 (AAQ87023) VDAC3.1, complete
Length = 1175
Score = 421 bits (1082), Expect = e-119
Identities = 211/261 (80%), Positives = 232/261 (88%)
Frame = +2
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDYNFD KF+LSIPSSTGLGL ATGLK+D+ F+GD+NTLYKSGN+TVDVKV+T SNV
Sbjct: 197 LLYKDYNFDQKFNLSIPSSTGLGLIATGLKKDQIFIGDINTLYKSGNLTVDVKVDTYSNV 376
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
STKVTLND+ KKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNP+P+LELS AI
Sbjct: 377 STKVTLNDIF--RCKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPSPQLELSTAI 550
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GSKD+ MGAEVGF+TTSASF+ YNAGI FN+PDFSA LML DKG+SLKASYIH+VDRPDG
Sbjct: 551 GSKDLCMGAEVGFNTTSASFTKYNAGITFNRPDFSAALMLVDKGKSLKASYIHHVDRPDG 730
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
TVAAE+ H+ SS ENRFT G+SQ IDP TVLKTRFSDDGKAAF CQRAWRP SLITLSA
Sbjct: 731 FTVAAEINHRFSSFENRFTIGSSQLIDPNTVLKTRFSDDGKAAFLCQRAWRPKSLITLSA 910
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
EYDS KI GS K GLAL+L+
Sbjct: 911 EYDSTKIFGSSTKLGLALALQ 973
Score = 26.9 bits (58), Expect = 3.5
Identities = 15/34 (44%), Positives = 21/34 (61%)
Frame = -1
Query: 198 FTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWR 231
FTF T +P VL+T ++DD K+ +CQR R
Sbjct: 1094 FTFMTRL*NNPSWVLETMYNDDCKS--ECQRIMR 999
>TC7918 UP|AAQ87019 (AAQ87019) VDAC1.1, complete
Length = 1601
Score = 226 bits (576), Expect = 3e-60
Identities = 117/261 (44%), Positives = 166/261 (62%)
Frame = +2
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY+ D KF+++ S TG+ +T++G K+ + FV D+NT K+ N+T D+KV+TDSN+
Sbjct: 131 LLFKDYHSDQKFTITTFSPTGVAITSSGTKKGELFVADVNTQLKNKNITTDIKVDTDSNL 310
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T + +ND + K SF +PD +SGK+++QYLH +A I SSIGL P + S I
Sbjct: 311 FTTIAVNDPA--PGLKAIFSFKVPDQRSGKVELQYLHDYAGISSSIGLTANPIVNFSGVI 484
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++GA++ FDT + NAG++F K D A L L DKG +L ASY H V
Sbjct: 485 GTNVLALGADLTFDTKIGELTKSNAGLSFTKDDLIAALTLNDKGDALNASYYHVVSPLTN 664
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE+ H+ S++EN T GT ++DP T LK R ++ GKA+ Q WRP S T+S
Sbjct: 665 TAVGAEVTHRFSTNENTLTLGTQHALDPLTTLKARVNNFGKASALIQHEWRPKSFFTISG 844
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GLAL+LK
Sbjct: 845 EVDTKAIEKS-AKVGLALALK 904
>TC8300 UP|AAQ87020 (AAQ87020) VDAC1.2, complete
Length = 1293
Score = 217 bits (552), Expect = 2e-57
Identities = 109/261 (41%), Positives = 166/261 (62%)
Frame = +3
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY+ D KF+++ S TG+ +T++G+K+ + FV D+NT K+ N+T D+KV+TDSN+
Sbjct: 138 LLFKDYHSDQKFTITTYSPTGVAITSSGIKKGELFVADVNTQLKNKNITTDLKVDTDSNL 317
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T VT+++ + K SF +PD +SGK+++QYLH +A I +S+GL P P + S +
Sbjct: 318 FTTVTVDEPA--PGLKAIFSFRVPDQRSGKVELQYLHDYAGISTSVGLTPNPIVNFSGVV 491
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++G ++ FDT + +NAG+ F K D A+L + DKG +L ASY H V+
Sbjct: 492 GTNALAVGTDLSFDTKIGELTKFNAGLNFTKADLIASLTVNDKGDALNASYYHIVNPLTN 671
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
AE+ H+ S++EN T GT ++DP T +K R ++ GKA Q WR S T+S
Sbjct: 672 TAFGAEVTHRFSTNENTLTLGTQHALDPLTSVKARVNNLGKANALIQHEWRAKSFFTISG 851
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GL L+LK
Sbjct: 852 EVDTKAIEKS-AKIGLGLALK 911
>TC8287 UP|AAQ87022 (AAQ87022) VDAC2.1, complete
Length = 1194
Score = 216 bits (549), Expect = 4e-57
Identities = 113/261 (43%), Positives = 170/261 (64%)
Frame = +1
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL KDY+ D K ++S S+ G+ LT+T +K+ VGD+ LYK N +DVK++T S +
Sbjct: 142 LLTKDYSSDQKLTVSSYSTAGVALTSTAVKKGGLSVGDVAALYKYKNTLIDVKLDTASII 321
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
ST +T D L S K SF +PD+ SGKL+VQY H HA + ++ LN +P +++SA
Sbjct: 322 STTLTFTD--LLPSTKAIASFKLPDYNSGKLEVQYFHDHATLTAAAALNQSPIIDVSATF 495
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ I++GAE G+DT+S F+ Y AGI+ KPD SA+++L DKG S+KASY+H++D+
Sbjct: 496 GTPVIAVGAEAGYDTSSGGFTKYTAGISVTKPDASASIILGDKGDSIKASYLHHLDQLKK 675
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
AE+ K S++EN FT G S +ID T ++ R ++ GK Q P S++T+S+
Sbjct: 676 SAAVAEITRKFSTNENTFTVGGSFAIDHLTQVRARLNNHGKLGALLQHEIIPKSVLTISS 855
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K + +P +FGLA++LK
Sbjct: 856 EVDTKALDKNP-RFGLAIALK 915
>TC8223 UP|AAQ87021 (AAQ87021) VDAC1.3, complete
Length = 1261
Score = 213 bits (542), Expect = 3e-56
Identities = 111/261 (42%), Positives = 167/261 (63%)
Frame = +2
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY DHKF+++ +S G+ +T+TG+K+ + ++ D++T K+ N+T DVKV+T+SN+
Sbjct: 80 LLFKDYQNDHKFTITTYTSAGVEITSTGVKKGEIYLADVSTKLKNKNITTDVKVDTNSNL 259
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ + K SF PD KSGK+++QY H +A I +SIGL +P + S I
Sbjct: 260 HTTITVDEPA--PGLKTIFSFIFPDQKSGKVELQYQHEYAGISTSIGLTASPIVNFSGII 433
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++G ++ FDT S +F+ YNAG+ D A+L L DKG SL ASY H V
Sbjct: 434 GNNLVAVGTDLSFDTASGNFTKYNAGLNVTHADLIASLTLNDKGDSLNASYYHVVSPLTN 613
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL+H SS+EN T GT ++DP T+LK R ++ G+A+ Q W P + ++L
Sbjct: 614 TAVGAELSHSFSSNENILTIGTQHALDPITLLKARVNNYGRASALIQHEWNPRTRVSLVG 793
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+ I S AK GLAL+LK
Sbjct: 794 EVDTGAIEKS-AKVGLALALK 853
>AV772563
Length = 477
Score = 40.0 bits (92), Expect = 4e-04
Identities = 19/48 (39%), Positives = 26/48 (53%)
Frame = -3
Query: 189 HKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLI 236
H+ S EN T GT ++DP T +K R + G+A Q WR SL+
Sbjct: 475 HRFSIHENTLTLGTQHALDPLTSVKARVNHLGQANAFIQHEWRAQSLL 332
>TC15910 similar to UP|Q9AR64 (Q9AR64) Urease accessory protein G, partial
(88%)
Length = 1061
Score = 28.9 bits (63), Expect = 0.93
Identities = 37/141 (26%), Positives = 57/141 (40%)
Frame = +2
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
L+ +D+N + F++ I G G TA L +F L Y VT D+ D
Sbjct: 236 LINRDFN-ERAFTIGIGGPVGTGKTALMLALCEF----LRDNYSLAAVTNDIFTKEDGEF 400
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
+ + +P+ + ++ PHAAI I +N P ELS +
Sbjct: 401 LVR----------------NKALPEERIRAVETGGC-PHAAIREDISINLGPLEELS-NL 526
Query: 121 GSKDISMGAEVGFDTTSASFS 141
DI + E G D +A+FS
Sbjct: 527 YKTDILL-CESGGDNLAANFS 586
>BF177857
Length = 384
Score = 27.3 bits (59), Expect = 2.7
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = -1
Query: 21 GLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDS 58
GLGL GL G+L L + N+ V VNTD+
Sbjct: 276 GLGLVGRGLGDQLLEAGNLANLLEEENIAWLVSVNTDA 163
>AV779028
Length = 522
Score = 27.3 bits (59), Expect = 2.7
Identities = 13/28 (46%), Positives = 14/28 (49%)
Frame = -2
Query: 17 PSSTGLGLTATGLKRDKFFVGDLNTLYK 44
PS G G+T G D FFVG T K
Sbjct: 341 PSQYGNGITENGFSSDPFFVGSPRTRAK 258
>BP046590
Length = 498
Score = 26.9 bits (58), Expect = 3.5
Identities = 16/53 (30%), Positives = 23/53 (43%)
Frame = +2
Query: 62 TKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKL 114
T + N ++ KK L+ H +G D +Y HP D + L P P L
Sbjct: 65 TNILYNKSTI*QKKKKNLT-----HHNGNQDNEYKHPSTHNDFDVSLFPPPTL 208
>AI967315
Length = 1308
Score = 26.6 bits (57), Expect = 4.6
Identities = 9/17 (52%), Positives = 13/17 (75%)
Frame = +1
Query: 5 DYNFDHKFSLSIPSSTG 21
+Y +D FS+S+P STG
Sbjct: 1045 EYEYDSSFSMSLPDSTG 1095
>TC16215 weakly similar to UP|Q9FI12 (Q9FI12) Serine proteinase, partial
(15%)
Length = 671
Score = 26.6 bits (57), Expect = 4.6
Identities = 13/36 (36%), Positives = 21/36 (58%)
Frame = +1
Query: 45 SGNVTVDVKVNTDSNVSTKVTLNDVSLSHSKKVALS 80
+GN T V + S VS KVT + +++H ++ LS
Sbjct: 190 AGNETYSVGWDAPSGVSVKVTPSHFTIAHGERKVLS 297
>BP058190
Length = 607
Score = 26.6 bits (57), Expect = 4.6
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 2/37 (5%)
Frame = +3
Query: 136 TSASFSTYNAGIAFNKPDFSA--TLMLADKGQSLKAS 170
++ASFSTY AG + P F+A L L +K + +K S
Sbjct: 396 STASFSTYLAGNSSISPIFAAKFRLKLIEKQEGIKVS 506
>TC14747
Length = 447
Score = 26.2 bits (56), Expect = 6.1
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 9/53 (16%)
Frame = -1
Query: 101 AIDSSIGL-------NPAPKLELSAAIGSK--DISMGAEVGFDTTSASFSTYN 144
A+DS+ GL + +PK + S + + D+S+ A G SASF+T N
Sbjct: 309 AVDSTKGLLMHSKSLHSSPKSKPSTILSTSEYDVSVAASEGSSVLSASFATRN 151
>AV780707
Length = 429
Score = 25.8 bits (55), Expect = 7.9
Identities = 17/54 (31%), Positives = 27/54 (49%)
Frame = +1
Query: 18 SSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNVSTKVTLNDVSL 71
SS G+G T G K+ VGD + VKVN+ +++ T++ + SL
Sbjct: 190 SSLGMGFTFLGSYPHKYAVGD----------ELSVKVNSLTSIDTEMPFSYYSL 321
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,731,356
Number of Sequences: 28460
Number of extensions: 43472
Number of successful extensions: 174
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 169
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 169
length of query: 261
length of database: 4,897,600
effective HSP length: 88
effective length of query: 173
effective length of database: 2,393,120
effective search space: 414009760
effective search space used: 414009760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC142506.2


