

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142498.7 + phase: 0
(1705 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
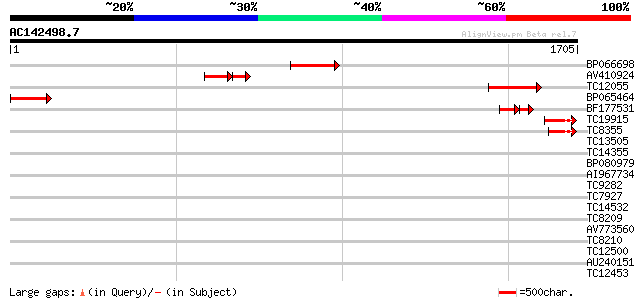
Score E
Sequences producing significant alignments: (bits) Value
BP066698 287 1e-77
AV410924 175 7e-71
TC12055 homologue to UP|Q8L3R8 (Q8L3R8) AT3g08530/T8G24_1, parti... 238 8e-63
BP065464 204 7e-53
BF177531 120 3e-47
TC19915 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, pa... 164 1e-40
TC8355 similar to UP|Q39834 (Q39834) Clathrin heavy chain, parti... 128 9e-30
TC13505 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, par... 39 0.007
TC14355 35 0.076
BP080979 35 0.099
AI967734 35 0.13
TC9282 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxy... 35 0.13
TC7927 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like... 33 0.38
TC14532 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor ... 33 0.49
TC8209 homologue to PIR|T10586|T10586 small nuclear ribonucleopr... 32 0.64
AV773560 32 0.64
TC8210 homologue to PIR|T10586|T10586 small nuclear ribonucleopr... 32 0.64
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 32 0.84
AU240151 32 1.1
TC12453 weakly similar to GB|BAA88270.1|6573289|AB008023 RXW8 {A... 31 1.9
>BP066698
Length = 442
Score = 287 bits (734), Expect = 1e-77
Identities = 141/147 (95%), Positives = 142/147 (95%)
Frame = +1
Query: 845 KGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNN 904
KGLILSVRSLLPVEPLV ECEKRNRLRLL+QFLEHLVSEGSQD HVHNALGKIIIDSNNN
Sbjct: 1 KGLILSVRSLLPVEPLVEECEKRNRLRLLSQFLEHLVSEGSQDAHVHNALGKIIIDSNNN 180
Query: 905 PEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVE 964
PEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRG CDDELINVTNKNSLFKLQARYVVE
Sbjct: 181 PEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGQCDDELINVTNKNSLFKLQARYVVE 360
Query: 965 RMDADLWEKVLNPDNTYRRQLIDQVVS 991
RMD DLWEKVLNPDN YRRQLIDQVVS
Sbjct: 361 RMDGDLWEKVLNPDNAYRRQLIDQVVS 441
>AV410924
Length = 421
Score = 175 bits (444), Expect(2) = 7e-71
Identities = 85/86 (98%), Positives = 86/86 (99%)
Frame = +3
Query: 585 LKPNLPEHGFLQTKVLEINLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQH 644
LKPNLPEHGFLQTKVLEINLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQH
Sbjct: 3 LKPNLPEHGFLQTKVLEINLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQH 182
Query: 645 YTELPDIKRVIVNTHAIEPQALVEFF 670
YTELPDIKRVIVNTHAIEPQ+LVEFF
Sbjct: 183 YTELPDIKRVIVNTHAIEPQSLVEFF 260
Score = 111 bits (277), Expect(2) = 7e-71
Identities = 52/55 (94%), Positives = 54/55 (97%)
Frame = +1
Query: 669 FFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSY 723
FFGTLS+EWALECMKDLLL NLRGNLQIIVQVAKEYCEQLGVDACIK+FEQFRSY
Sbjct: 256 FFGTLSQEWALECMKDLLLANLRGNLQIIVQVAKEYCEQLGVDACIKLFEQFRSY 420
>TC12055 homologue to UP|Q8L3R8 (Q8L3R8) AT3g08530/T8G24_1, partial (22%)
Length = 483
Score = 238 bits (606), Expect = 8e-63
Identities = 125/160 (78%), Positives = 131/160 (81%), Gaps = 1/160 (0%)
Frame = +1
Query: 1440 VLALRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESI 1499
VLALRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESI
Sbjct: 1 VLALRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESI 180
Query: 1500 DLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQS 1559
DLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYK +
Sbjct: 181 DLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKACNGRQPHN 360
Query: 1560 -GERELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVAL 1598
L L + + ++ CLFVCYDLI D+AL
Sbjct: 361 LASVNLLRNCLFISLIRERRNALPRCLFVCYDLIGADIAL 480
>BP065464
Length = 430
Score = 204 bits (520), Expect = 7e-53
Identities = 98/122 (80%), Positives = 109/122 (89%)
Frame = +3
Query: 4 AANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPNQPL 63
AANAPI M+E LTL S+GINPQ ITFTHVTMES++YICVRETAPQNSVV++DMN+ QPL
Sbjct: 57 AANAPISMKEVLTLGSVGINPQFITFTHVTMESEQYICVRETAPQNSVVMIDMNIAMQPL 236
Query: 64 RRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISP 123
RRPITADSALMNPN RILALKAQ+ GTTQDHLQ+FNIE K KMKSY M +QVVFWKWI P
Sbjct: 237 RRPITADSALMNPNXRILALKAQVPGTTQDHLQVFNIETKTKMKSYPMTQQVVFWKWIXP 416
Query: 124 KL 125
K+
Sbjct: 417 KI 422
>BF177531
Length = 324
Score = 120 bits (300), Expect(2) = 3e-47
Identities = 59/61 (96%), Positives = 61/61 (99%)
Frame = +2
Query: 1472 SNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELLEMRRVAAYIYK 1531
SNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELLE+RRVAAYIYK
Sbjct: 14 SNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELLEIRRVAAYIYK 193
Query: 1532 K 1532
+
Sbjct: 194 Q 196
Score = 87.4 bits (215), Expect(2) = 3e-47
Identities = 43/43 (100%), Positives = 43/43 (100%)
Frame = +3
Query: 1532 KAGRWKQSIALSKKDNLYKDAMETASQSGERELAEELLVYFID 1574
KAGRWKQSIALSKKDNLYKDAMETASQSGERELAEELLVYFID
Sbjct: 195 KAGRWKQSIALSKKDNLYKDAMETASQSGERELAEELLVYFID 323
>TC19915 homologue to UP|Q39834 (Q39834) Clathrin heavy chain, partial (5%)
Length = 447
Score = 164 bits (415), Expect = 1e-40
Identities = 82/96 (85%), Positives = 85/96 (88%)
Frame = +3
Query: 1609 FAFPYLLQFIREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPP 1668
FAFPY+LQ +REYTGKVDELVK KIE+Q EVKAKEQEEKEVI QQNMYAQLLPLALPAPP
Sbjct: 3 FAFPYVLQLLREYTGKVDELVKDKIEAQKEVKAKEQEEKEVIQQQNMYAQLLPLALPAPP 182
Query: 1669 MPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
MP GMGGGYAPPPPM GGMGMPPMPPFGMP G
Sbjct: 183 MP----GMGGGYAPPPPM-GGMGMPPMPPFGMPPMG 275
Score = 30.8 bits (68), Expect = 1.9
Identities = 16/32 (50%), Positives = 17/32 (53%)
Frame = -3
Query: 1668 PMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFG 1699
PM GM G GG PP GGG PP+P G
Sbjct: 274 PMGGMPKGGIGGIPIPPIGGGGA*PPPIPGIG 179
>TC8355 similar to UP|Q39834 (Q39834) Clathrin heavy chain, partial (4%)
Length = 578
Score = 128 bits (321), Expect = 9e-30
Identities = 67/87 (77%), Positives = 74/87 (85%), Gaps = 1/87 (1%)
Frame = +2
Query: 1619 REYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGG 1678
REYTGKVDELVK KIE+ E KAKE+EEK+V++QQNMYAQLLPLALPAPPMP GMGG
Sbjct: 2 REYTGKVDELVKDKIEAPIEEKAKEKEEKDVVSQQNMYAQLLPLALPAPPMP----GMGG 169
Query: 1679 GYAPPPPMGGGMGMPPMPPFGM-PMGG 1704
GY PPPPM GG+GMP PP+GM PMGG
Sbjct: 170 GYGPPPPM-GGLGMP--PPYGMPPMGG 241
Score = 32.0 bits (71), Expect = 0.84
Identities = 16/28 (57%), Positives = 16/28 (57%)
Frame = -3
Query: 1670 PGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P MGG GG P PPMGGG PP P
Sbjct: 240 PPMGGMP*GGGIPSPPMGGGGP*PPPIP 157
Score = 31.2 bits (69), Expect = 1.4
Identities = 17/33 (51%), Positives = 18/33 (54%)
Frame = -3
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFG 1699
PPM GM G GG PP GGG PP+P G
Sbjct: 240 PPMGGMP*G--GGIPSPPMGGGGP*PPPIPGIG 148
>TC13505 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (7%)
Length = 359
Score = 38.9 bits (89), Expect = 0.007
Identities = 18/40 (45%), Positives = 19/40 (47%)
Frame = +1
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P PP P GGG GG Y PPP + P PP GG
Sbjct: 223 PPPPSPPSGGGGGGSYYSPPPPSQYVYSSPPPPASTGXGG 342
>TC14355
Length = 687
Score = 35.4 bits (80), Expect = 0.076
Identities = 21/44 (47%), Positives = 22/44 (49%), Gaps = 10/44 (22%)
Frame = +2
Query: 1670 PGMGGGMGGGYAPPPPMGGGMGMPPM----------PPFGMPMG 1703
PG+G GMGG P MG GMGM M PP GMP G
Sbjct: 68 PGVGMGMGGYGGMNPSMGMGMGMGGMGMGQGYQVQQPPAGMPPG 199
>BP080979
Length = 352
Score = 35.0 bits (79), Expect = 0.099
Identities = 19/59 (32%), Positives = 25/59 (42%), Gaps = 3/59 (5%)
Frame = +3
Query: 1643 EQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGG---MGGGYAPPPPMGGGMGMPPMPPF 1698
EQ + + LP P PP G GG + PP P+G +PP+PPF
Sbjct: 159 EQSQSTQSVGKRSNGHRLPXPTPPPPPQGPQGGRQILPPRRRPPRPLGNQRPLPPLPPF 335
>AI967734
Length = 465
Score = 34.7 bits (78), Expect = 0.13
Identities = 18/39 (46%), Positives = 21/39 (53%)
Frame = +2
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGGY 1705
PP PG +G PPP GG PP PP+ +P GGY
Sbjct: 194 PPRPGPHPAIGFPQPVPPPGAGGPSAPP-PPY-LPHGGY 304
>TC9282 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxykinase ,
partial (44%)
Length = 1236
Score = 34.7 bits (78), Expect = 0.13
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 16/105 (15%)
Frame = +3
Query: 734 LSSSEDPDIHFKYIEAAAKTGQIKE---VERVTRESSFYDPEKTKN--------FLMEAK 782
LS ++PDI A + G + E + TRE + D T+N ++ AK
Sbjct: 105 LSREKEPDIW-----NAIRFGTVLENVVFDEHTREVDYSDKSVTENTRAAYPIEYIPNAK 269
Query: 783 LPDARP-----LINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKV 822
LP P ++ CD FG +P ++ M +I GY V
Sbjct: 270 LPCVGPHPTNVILLACDAFGVLPPVSKLNLAQTMYHFISGYTALV 404
>TC7927 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like, partial
(56%)
Length = 1116
Score = 33.1 bits (74), Expect = 0.38
Identities = 31/100 (31%), Positives = 42/100 (42%), Gaps = 14/100 (14%)
Frame = +1
Query: 1618 IREYTGKVDELVKHKIESQNEVKAKEQEEKEVIA------QQNMYAQLLPLALPAPPMPG 1671
I E+ K + L K + E + E + + + + A Q+ + P +P PG
Sbjct: 271 IEEHRRKYERLHKEREEKRTERERQRRRAEAQAAYEKAKKQEQSSSSRNPGGMPGE-FPG 447
Query: 1672 MGGGMGGGYAPPPPMGGGM-------GMP-PMPPFGMPMG 1703
M GG GG P GGGM GMP P GMP G
Sbjct: 448 MPGGFPGGGMPGGFPGGGMPGGFPGGGMPGGFPGGGMPGG 567
Score = 29.6 bits (65), Expect = 4.2
Identities = 20/43 (46%), Positives = 20/43 (46%), Gaps = 3/43 (6%)
Frame = -2
Query: 1665 PAPPMPGMGGGMGGGYAPP--PPMGGGMGMPPM-PPFGMPMGG 1704
P P PG GM PP PP G GMPP P GMP G
Sbjct: 644 PGMPPPGNPPGMPPPGNPPGMPPPGNPPGMPPPGKPPGMPPPG 516
>TC14532 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor SCL30a, 30a
kD, partial (71%)
Length = 1073
Score = 32.7 bits (73), Expect = 0.49
Identities = 19/45 (42%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Frame = +3
Query: 1652 QQNMYAQLLPLALPAPPMPGMG--GGMGGGYAPPPPMGGGMGMPP 1694
Q+NM + + P PP G G GG GGG +P P GG G P
Sbjct: 21 QRNMRGRSYTPSPPPPPPRGYGRRGGGGGGRSPSPRGGGRYGPRP 155
>TC8209 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (19%)
Length = 635
Score = 32.3 bits (72), Expect = 0.64
Identities = 18/34 (52%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Frame = +3
Query: 1665 PAPPMPGMGGGMGGG--YAPPPPMGGGMGMPPMP 1696
P PP PGM GG YAPP P GMPP P
Sbjct: 129 PPPPRPGMPPPPGGAPVYAPPRP-----GMPPPP 215
Score = 32.0 bits (71), Expect = 0.84
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = +3
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPM 1702
P PP PGM G PPPP GMPP PP G P+
Sbjct: 99 PPPPRPGMPG-------PPPPR---PGMPP-PPGGAPV 179
Score = 28.9 bits (63), Expect = 7.1
Identities = 18/35 (51%), Positives = 18/35 (51%)
Frame = +3
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
PP PG M G PPPP GM PP P GMP
Sbjct: 66 PPPPGQ---MMRG--PPPPPRPGMPGPPPPRPGMP 155
>AV773560
Length = 438
Score = 32.3 bits (72), Expect = 0.64
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = -3
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPM 1702
P PP PGM G PPPP GMPP PP G P+
Sbjct: 409 PPPPRPGMPG-------PPPPR---PGMPP-PPAGAPV 329
>TC8210 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (16%)
Length = 698
Score = 32.3 bits (72), Expect = 0.64
Identities = 18/34 (52%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Frame = +2
Query: 1665 PAPPMPGMGGGMGGG--YAPPPPMGGGMGMPPMP 1696
P PP PGM GG YAPP P GMPP P
Sbjct: 167 PPPPRPGMPPPPGGAPVYAPPRP-----GMPPPP 253
Score = 32.0 bits (71), Expect = 0.84
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = +2
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPM 1702
P PP PGM G PPPP GMPP PP G P+
Sbjct: 137 PPPPRPGMPG-------PPPPR---PGMPP-PPGGAPV 217
Score = 28.9 bits (63), Expect = 7.1
Identities = 18/35 (51%), Positives = 18/35 (51%)
Frame = +2
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
PP PG M G PPPP GM PP P GMP
Sbjct: 104 PPPPGQM--MRG---PPPPPRPGMPGPPPPRPGMP 193
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 32.0 bits (71), Expect = 0.84
Identities = 16/40 (40%), Positives = 19/40 (47%)
Frame = +1
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGM 1700
P +LP+PP P PPPP G +PP PP M
Sbjct: 292 PCSLPSPPQP----------PPPPPKYGVTTIPPSPPLLM 381
>AU240151
Length = 286
Score = 31.6 bits (70), Expect = 1.1
Identities = 20/41 (48%), Positives = 20/41 (48%), Gaps = 2/41 (4%)
Frame = -2
Query: 1665 PAPPMPG-MGGGMGGGYAPPP-PMGGGMGMPPMPPFGMPMG 1703
P PP PG G G G PPP P G G PP PP P G
Sbjct: 237 PGPPGPGPQPGPPGPGPQPPPGPPGPGPQPPPGPPPPGPPG 115
Score = 29.3 bits (64), Expect = 5.4
Identities = 19/40 (47%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Frame = +2
Query: 1667 PPMPGMGGGMGGGYAPPP--PMGGGMGMPPMPPFGMPMGG 1704
P PG GGG GGG+ P P P GG P P +G GG
Sbjct: 116 PGGPG-GGGPGGGWGPGPGGPGGGWGPGPGGPGWGPGPGG 232
>TC12453 weakly similar to GB|BAA88270.1|6573289|AB008023 RXW8 {Arabidopsis
thaliana;} , partial (10%)
Length = 528
Score = 30.8 bits (68), Expect = 1.9
Identities = 16/39 (41%), Positives = 22/39 (56%)
Frame = -2
Query: 180 GSPERPQLVKGNMQLFSVEQQRSQALEAHAASFAQFKVP 218
G+PE P +K +QLFS E+ S A+SF+ VP
Sbjct: 488 GAPEHPHCIKLGLQLFSSEESLSTLHICPASSFSFCTVP 372
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,556,078
Number of Sequences: 28460
Number of extensions: 346164
Number of successful extensions: 2170
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1922
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2092
length of query: 1705
length of database: 4,897,600
effective HSP length: 103
effective length of query: 1602
effective length of database: 1,966,220
effective search space: 3149884440
effective search space used: 3149884440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC142498.7


