

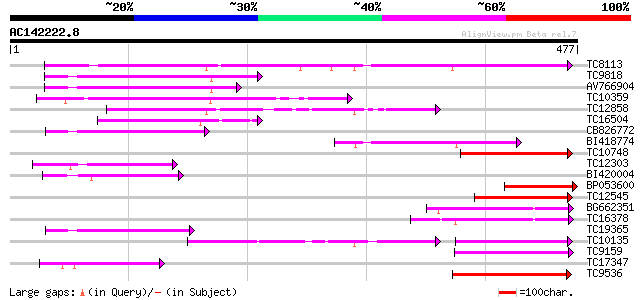
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.8 - phase: 0
(477 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 199 6e-52
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 124 3e-29
AV766904 103 6e-23
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 96 2e-20
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 96 2e-20
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 90 9e-19
CB826772 89 2e-18
BI418774 80 6e-16
TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 80 9e-16
TC12303 similar to GB|CAB10424.1|2245004|ATFCA6 membrane transpo... 79 2e-15
BI420004 78 3e-15
BP053600 77 5e-15
TC12545 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 77 6e-15
BG662351 76 1e-14
TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22... 72 3e-13
TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter l... 70 6e-13
TC10135 70 6e-13
TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose tr... 70 6e-13
TC17347 similar to UP|Q9FMX3 (Q9FMX3) Monosaccharide transporter... 70 1e-12
TC9536 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, parti... 67 5e-12
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 199 bits (507), Expect = 6e-52
Identities = 135/463 (29%), Positives = 233/463 (50%), Gaps = 19/463 (4%)
Frame = +2
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
++ GYD G S A + I+ T ++ A + + AL+G + A +D++
Sbjct: 152 IISGYDTGVMSGALLFIKEDIGISDTQQEVLAGILNIC-------ALVGCLAAGKTSDYI 310
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR + +A++++LVGA+ + PNF +L+ GR V G+G+G A+ AP+Y AE + R
Sbjct: 311 GRRYTIFLASILFLVGAVFMGYGPNFAILMFGRCVCGLGVGFALTTAPVYSAELSSASTR 490
Query: 150 GQLVSLKEFFIVIGI----VAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
G L SL E I +GI ++ Y LG L + GWR M G+++ ++ + G+ +P S
Sbjct: 491 GFLTSLPEVCIGLGIFIGYISNYFLGKLAL--TLGWRLMLGLAAIPSLGLALGILTMPES 664
Query: 206 PRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSA--PQQVDEIMAEFSYLGEENDV 263
PRW++++ L+ + +L+ R SA ++ ++ + +
Sbjct: 665 PRWLVMQGRLGCAKKVLLEVSNTTEEAELRFRDIIVSAGFDEKCNDEFVKQPQKSHHGEG 844
Query: 264 TLGEMF---RGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADA 317
E+F R+ L+ + G+ F+ I+ V +GP R G +
Sbjct: 845 VWKELFLRPTPPVRRMLIAAVGIHFFEHATGIEAVMLYGP------RIFKKAGVTTKDRL 1006
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN------ 371
+I G+ K+ ++ ++DR+GRR LL V+G++ L +LG ++
Sbjct: 1007LLATIGTGLTKITFLTISTFLLDRVGRRRLLQISVAGMIFGLTILGFSLTMVEYSSEKLV 1186
Query: 372 -AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAF 430
A L++V YV + + P+ W+ +EIFPLRLR +G SI V VN NA ++ +F
Sbjct: 1187WALSLSIVATYTYVAFFNVGLAPVTWVYSSEIFPLRLRAQGNSIGVAVNRGMNAAISMSF 1366
Query: 431 SPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+ + G F++F+ ++V + VF YF +PETKG LEE+E
Sbjct: 1367ISIYKAITIGGAFFLFAGMSVVAWVFFYFCLPETKGKALEEME 1495
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 124 bits (311), Expect = 3e-29
Identities = 72/185 (38%), Positives = 104/185 (55%), Gaps = 2/185 (1%)
Frame = +2
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A I I+ + V+I +L +LIGS LA +D++
Sbjct: 272 ILLGYDIGVMSGAAIYIKRD-------LKVSDVKIEILLGIINLYSLIGSGLAGRTSDWI 430
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR ++ A ++ VGAL+ F+PN+ L+ GR + GIGIG A+ AP+Y AE +P R
Sbjct: 431 GRRYTIVFAGAIFFVGALLMGFSPNYWFLMFGRFIAGIGIGYALMIAPVYTAEVSPASSR 610
Query: 150 GQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E FI GI+ GY + GWR M G+ + +VI+G G+ +P SPR
Sbjct: 611 GFLTSFPEVFINGGILLGYISNFAFSKLSLKVGWRMMLGVGALPSVILGVGVLAMPESPR 790
Query: 208 WILLR 212
W+++R
Sbjct: 791 WLVMR 805
Score = 35.0 bits (79), Expect = 0.027
Identities = 38/159 (23%), Positives = 66/159 (40%), Gaps = 11/159 (6%)
Frame = +2
Query: 307 NPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY 366
N F+ A A+ SILLG +M+G A+ + L + + + GI+ L+GS
Sbjct: 227 NKYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDVKIEILLGIINLYSLIGSGL 406
Query: 367 I-----FLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFA 421
++ + G + +VG + F P W + +F + G G+ A+++
Sbjct: 407 AGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYWFL---MFGRFIAGIGIGYALMIAPV 577
Query: 422 ANALVT------FAFSPLKDLLGAGILFYIFSAIAVASL 454
A V+ F S + + GIL S A + L
Sbjct: 578 YTAEVSPASSRGFLTSFPEVFINGGILLGYISNFAFSKL 694
>AV766904
Length = 611
Score = 103 bits (257), Expect = 6e-23
Identities = 61/168 (36%), Positives = 93/168 (55%), Gaps = 2/168 (1%)
Frame = +3
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A I I+ + V+I +L+ + IGS +A +D++
Sbjct: 126 ILLGYDIGVMSGAVIYIKRD-------LKISDVQIEILSGIMNIYSPIGSYIAGRTSDWI 284
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A L++ VGA++ F+PN+ L+ GR V G+GIG A AP+Y +E +PT R
Sbjct: 285 GRRYTIVLAGLIFFVGAILMGFSPNYAFLMFGRFVAGVGIGFAFLIAPVYTSEISPTLSR 464
Query: 150 GQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIM 195
G L SL E F+ GI+ GY + GWR M GI + ++ +
Sbjct: 465 GFLTSLPEVFLNAGILIGYISNYTFSKLPLRLGWRLMLGIGAIPSIFL 608
Score = 32.0 bits (71), Expect = 0.23
Identities = 29/118 (24%), Positives = 51/118 (42%), Gaps = 6/118 (5%)
Frame = +3
Query: 307 NPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY 366
N + A A+ SILLG +M+G + + L + + +SGI+ +GSY
Sbjct: 81 NKFALACAILASMTSILLGYDIGVMSGAVIYIKRDLKISDVQIEILSGIMNIYSPIGSYI 260
Query: 367 I-----FLDNAAVLAVVGLLLYVGCYQISFGP-MGWLMIAEIFPLRLRGKGLSIAVLV 418
++ + + GL+ +VG + F P +LM + G G+ A L+
Sbjct: 261 AGRTSDWIGRRYTIVLAGLIFFVGAILMGFSPNYAFLMFGRF----VAGVGIGFAFLI 422
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 95.5 bits (236), Expect = 2e-20
Identities = 73/284 (25%), Positives = 128/284 (44%), Gaps = 18/284 (6%)
Frame = +2
Query: 23 LFPAFGGLLFGYDIGATSSATIS----------------IQSSSLSGITWYDLDAVEIGL 66
+ A GGL+FGYD+G + T ++ S YD ++ L
Sbjct: 224 ILAATGGLMFGYDVGVSGGVTSMPHFLHKFFPDVYKKTVLEKELDSNYCKYDNQGLQ--L 397
Query: 67 LTSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFG 126
TS L+ + A LGRR +L+A + ++ G + A N +L++GR++ G
Sbjct: 398 FTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFFIAGVVFNVTAQNLAMLIVGRILLG 577
Query: 127 IGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAG--YGLGSLLVDTVAGWRYM 184
G+G A A P++++E AP+ IRG L L + I IGI+ G+ + GWR
Sbjct: 578 CGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITIGILFANLINYGTNKIKGGWGWRLS 757
Query: 185 FGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAP 244
G++ AV++ G + +P ++ R ++G K IR ++ P
Sbjct: 758 LGLAGIPAVLLTVGALLVVDTPNSLIERGRTEEGKAVLKK---IRGTDNVE--------P 904
Query: 245 QQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ 288
+ ++ + E S + +E + + + R ++IS L +FQQ
Sbjct: 905 EFLE--LVEASRVAKEVKDPFRNLLKRRNRPQIIISIALQIFQQ 1030
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 95.5 bits (236), Expect = 2e-20
Identities = 85/288 (29%), Positives = 132/288 (45%), Gaps = 7/288 (2%)
Frame = +3
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
A I +G R ++V L ++ GAL A +L++GRL+ G GIG A + P+Y++
Sbjct: 12 ASTITRIMGMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIYLS 191
Query: 142 ETAPTPIRGQLVSLKEFFIVIGI----VAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGF 197
E AP RG L + I IGI + Y +L GWR G+ + AVI
Sbjct: 192 EMAPYKYRGGLNMCFQLSITIGIFVANLFNYYFAKIL--NGQGWRLSLGLGAVPAVIFVV 365
Query: 198 GMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYL 257
G LP SP ++ R + A + L +++G T D + D I A S
Sbjct: 366 GSLCLPDSPSSLVARG---------RHEAARQELVKIRGTT--DIEAELKDIITA--SEA 506
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLA 314
E + K R LV + + FQQ + +T + P L R I GF
Sbjct: 507 LENVKHPWKTLLERKYRPQLVFAVCIPFFQQFTGLNVITFYAPI---LFRTI---GFGPT 668
Query: 315 ADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLL 362
A + ++++G FK + T +++ VVD+ GRR L L G + ++I ++
Sbjct: 669 A-SLMSAVIIGSFKPVSTLISIFVVDKFGRRTLFLEGGAQMLICQIIM 809
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 89.7 bits (221), Expect = 9e-19
Identities = 49/143 (34%), Positives = 85/143 (59%), Gaps = 5/143 (3%)
Frame = +2
Query: 75 ALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMH 134
AL + A IAD++GRR +++ A+++LVG+++ + P++P+L+IG + GIG+G ++
Sbjct: 236 ALPACMTAGRIADYIGRRYTIILTAIIFLVGSVLMGYGPSYPILMIGLSISGIGLGFSLI 415
Query: 135 AAPMYIAETAPTPIRGQLVSLKEFF----IVIGIVAGYGLGSLLVDTVAGWRYMFGISS- 189
AP+Y AE +P RG L S E I +G V+ Y G L + GWR M + +
Sbjct: 416 VAPVYCAEISPPSHRGFLTSFLEVSTNAGIALGYVSNYFFGKLSIR--LGWRLMLVVHAV 589
Query: 190 PVAVIMGFGMWWLPASPRWILLR 212
P ++ + W+ SPRW++++
Sbjct: 590 PSLALVALMLNWV-ESPRWLVMQ 655
>CB826772
Length = 490
Score = 88.6 bits (218), Expect = 2e-18
Identities = 45/138 (32%), Positives = 80/138 (57%)
Frame = +1
Query: 31 LFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLG 90
++GY++G + A + I+ + +++ L L S+ A I+D++G
Sbjct: 94 IYGYEVGVMTGALLFIKED-------LQISDLQLQFLVGIFHMCGLPASMAAGRISDYIG 252
Query: 91 RRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRG 150
RR +++A++ +L G+++T ++P++ +L+IG + GIG G + AP+Y AE +P RG
Sbjct: 253 RRYTIILASIAFLSGSILTGYSPSYLILMIGNFISGIGAGFTLIVAPLYCAEISPPSHRG 432
Query: 151 QLVSLKEFFIVIGIVAGY 168
L SL EF I IGI+ GY
Sbjct: 433 SLTSLTEFSINIGILLGY 486
>BI418774
Length = 554
Score = 80.5 bits (197), Expect = 6e-16
Identities = 50/167 (29%), Positives = 86/167 (50%), Gaps = 10/167 (5%)
Frame = +3
Query: 274 RKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLI 330
R+ L+ + G+ +FQQI + + + P R TG + ++ ++ LG+ + +
Sbjct: 63 RRILIAAIGVHVFQQICGIEGICIYSP------RVFEKTGITSKSNLLLATVGLGISQAV 224
Query: 331 MTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAV-------LAVVGLLLY 383
T ++ ++DR+GRR LLL G+V++L L + + +V + ++
Sbjct: 225 FTFISAFLLDRVGRRILLLISSGGVVVTLLGLSFCSFIMQQSKEEPLWGISFTIVAIYIF 404
Query: 384 VGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAF 430
V I GP+ W+ +EIFPLRLR +GL + VLVN AN V +F
Sbjct: 405 VAFVAIGIGPVTWVYSSEIFPLRLRAQGLGVCVLVNRLANVAVLTSF 545
>TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(21%)
Length = 550
Score = 79.7 bits (195), Expect = 9e-16
Identities = 36/94 (38%), Positives = 59/94 (62%)
Frame = +2
Query: 380 LLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGA 439
+L YV + I GP+ W+ +EIFPLRLR +G ++ V+VN + +++ F L +
Sbjct: 5 VLSYVATFSIGAGPITWVYSSEIFPLRLRAQGCAMGVVVNRVTSGVISMTFLSLSKGITI 184
Query: 440 GILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
G F++F IA+ +F Y ++PET+G TLE++E
Sbjct: 185 GGAFFLFGGIAICGWIFFYTMLPETRGKTLEDME 286
>TC12303 similar to GB|CAB10424.1|2245004|ATFCA6 membrane transporter like
protein {Arabidopsis thaliana;}, partial (25%)
Length = 545
Score = 79.0 bits (193), Expect = 2e-15
Identities = 47/125 (37%), Positives = 73/125 (57%), Gaps = 3/125 (2%)
Frame = +3
Query: 20 LRFLFPA-FGGLLFGYDIGATSSATISIQSS--SLSGITWYDLDAVEIGLLTSGSLYGAL 76
LR F A GGLLFGYD G S A + I+ ++ TW + S ++ GA+
Sbjct: 189 LRRAFSAGIGGLLFGYDTGVISGALLYIKDDFKAVDRKTWLQ------EAIVSTAIAGAI 350
Query: 77 IGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAA 136
IG+ + I D GRR +++A ++L+G++I A APN +L+ GR+ G G+G+A A+
Sbjct: 351 IGAAVGGWINDRFGRRVAIIIADTLFLIGSVIMAAAPNPAILLFGRVFVGFGVGMASMAS 530
Query: 137 PMYIA 141
P+YI+
Sbjct: 531 PLYIS 545
>BI420004
Length = 547
Score = 78.2 bits (191), Expect = 3e-15
Identities = 47/124 (37%), Positives = 71/124 (56%), Gaps = 5/124 (4%)
Frame = +1
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLL-----TSGSLYGALIGSVLA 82
GGLLFGYD G S A + I+ D + V L S +L GA+IG+
Sbjct: 199 GGLLFGYDTGVISGALLYIKD---------DFEEVRRSSLLQETIVSMALVGAIIGAATG 351
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
I D GR++ L A +++ +G+L+ A AP+ ++ GRL+ G+GIG+A AP+YIAE
Sbjct: 352 GWINDAFGRKKATLSADVVFTLGSLVMAAAPDAYFVIFGRLLVGLGIGVASVTAPVYIAE 531
Query: 143 TAPT 146
++P+
Sbjct: 532 SSPS 543
>BP053600
Length = 400
Score = 77.4 bits (189), Expect = 5e-15
Identities = 37/61 (60%), Positives = 51/61 (82%)
Frame = -2
Query: 417 LVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKC 476
L +FA+NA+VT AFSPLK+LLGA LF +F A+++ +L+F+ F VPETKGL+LEEIE++
Sbjct: 399 LTHFASNAVVTLAFSPLKELLGAENLFLLFGALSLVALLFVIFSVPETKGLSLEEIESQI 220
Query: 477 L 477
L
Sbjct: 219 L 217
>TC12545 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(18%)
Length = 482
Score = 77.0 bits (188), Expect = 6e-15
Identities = 35/82 (42%), Positives = 55/82 (66%)
Frame = +2
Query: 392 GPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAV 451
GP+ W+ +EIFPLRLR +G++I +VN +++ F L + + G F++F+ IAV
Sbjct: 8 GPITWVYSSEIFPLRLRAQGVAIGAVVNRLTRGVISMTFLSLSNAITIGGAFFLFAGIAV 187
Query: 452 ASLVFIYFIVPETKGLTLEEIE 473
+ +F Y ++PET+G TLEEIE
Sbjct: 188 CAWIFHYTLLPETRGQTLEEIE 253
>BG662351
Length = 407
Score = 76.3 bits (186), Expect = 1e-14
Identities = 43/126 (34%), Positives = 75/126 (59%), Gaps = 2/126 (1%)
Frame = +1
Query: 351 GVSGIVISL--FLLGSYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLR 408
G G +++ F L + I + LAV G+L+YVG + I G + W++++EIFP+ ++
Sbjct: 7 GTRGSILTAVAFYLKAQEISVGAVPGLAVTGILVYVGSFAIGMGAVPWVVMSEIFPVNIK 186
Query: 409 GKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLT 468
G+ S+A LVN+ L ++ F+ L G F +++AI ++ I +VPETKG +
Sbjct: 187 GQAGSLATLVNWFGAWLCSYTFNFLMSWSTYG-TFILYAAINALGILSIVVVVPETKGKS 363
Query: 469 LEEIEA 474
LE+++A
Sbjct: 364 LEQLQA 381
>TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;}, partial (31%)
Length = 738
Score = 71.6 bits (174), Expect = 3e-13
Identities = 46/148 (31%), Positives = 82/148 (55%), Gaps = 11/148 (7%)
Frame = +2
Query: 338 VVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAA-----------VLAVVGLLLYVGC 386
+VD+ GRR LLL S + +SL ++ S +L+ +++VVGL++ V
Sbjct: 11 LVDKSGRRLLLLLSSSIMTVSLVVV-SIAFYLEGTVSEDSHLYKILGIVSVVGLVVMVIG 187
Query: 387 YQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIF 446
+ + GP+ W++++EI P+ ++G S A + N+ + +T + L G F ++
Sbjct: 188 FSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGG-TFTMY 364
Query: 447 SAIAVASLVFIYFIVPETKGLTLEEIEA 474
+ +A ++VF VPETKG TLEEI++
Sbjct: 365 TVVAAFTVVFTALWVPETKGRTLEEIQS 448
>TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (24%)
Length = 441
Score = 70.5 bits (171), Expect = 6e-13
Identities = 39/125 (31%), Positives = 72/125 (57%)
Frame = +2
Query: 31 LFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLG 90
+FGY G + A + I+ ++ +++ LL AL ++A +D++G
Sbjct: 83 IFGYVTGVMAGALLFIKEE-------LGINDMQVQLLGGILNVCALPACMVAGRTSDYVG 241
Query: 91 RRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRG 150
RR ++++A++ L+G+++ + P+FP+L+IGR G+G+GLA+ AP+Y E + RG
Sbjct: 242 RRYTIILSAVILLLGSILMGYGPSFPILMIGRCTAGLGVGLALIIAPVYSTEISLPSNRG 421
Query: 151 QLVSL 155
L SL
Sbjct: 422 FLTSL 436
>TC10135
Length = 1501
Score = 70.5 bits (171), Expect = 6e-13
Identities = 35/98 (35%), Positives = 57/98 (57%)
Frame = +2
Query: 376 AVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKD 435
A+VGL LY+ + G + W++ +EI+P R RG IA + +N +V+ +F L
Sbjct: 893 ALVGLALYIITFSPGMGTVPWVINSEIYPFRYRGVCGGIASTTVWISNLIVSQSFLSLTQ 1072
Query: 436 LLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+G F +F +AV ++ F+ VPETKG+ +EE+E
Sbjct: 1073TIGTAWTFMMFGILAVVAIFFVIVFVPETKGVPMEEVE 1186
Score = 64.3 bits (155), Expect = 4e-11
Identities = 59/216 (27%), Positives = 94/216 (43%), Gaps = 3/216 (1%)
Frame = +2
Query: 150 GQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWI 209
G LVSL F I G Y + + WR+M G+++ A+I M LP SPRW
Sbjct: 2 GALVSLNSFLITGGQFLSYLINLAFTNAPGTWRWMLGVAAAPAIIQIVLMLSLPESPRW- 178
Query: 210 LLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMF 269
L R +++ LK + +S +++E S + + +TL +
Sbjct: 179 LYRKGREEESKSILKKIYAPEDVDAEIEALKESVESEIEE-----SKTSKISMMTL--LK 337
Query: 270 RGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGV 326
R+ L GL FQQ I TV + PT L GF+ A +S++
Sbjct: 338 TTTVRRGLYAGMGLQFFQQFVGINTVMYYSPTIVQL------AGFASNRTALLLSLITSG 499
Query: 327 FKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLL 362
+ +++ +D+ GR+ L L + G+V+SL LL
Sbjct: 500 LNAFGSILSIYFIDKTGRKKLALISLCGVVLSLALL 607
>TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(5%)
Length = 728
Score = 70.5 bits (171), Expect = 6e-13
Identities = 36/100 (36%), Positives = 58/100 (58%)
Frame = +3
Query: 375 LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLK 434
LA+ GL LY+ + G + W++ +EI+PLR RG IA + +N +V+ +F L
Sbjct: 117 LALGGLALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSLT 296
Query: 435 DLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
+G F +F IA+ + F+ VPETKG+ +EE+E+
Sbjct: 297 QTIGTAWTFMMFGIIAIIGIFFVLIFVPETKGVPMEEVES 416
>TC17347 similar to UP|Q9FMX3 (Q9FMX3) Monosaccharide transporter (STP11
protein), partial (30%)
Length = 663
Score = 69.7 bits (169), Expect = 1e-12
Identities = 44/119 (36%), Positives = 56/119 (46%), Gaps = 14/119 (11%)
Frame = +3
Query: 26 AFGGLLFGYDIGATSSAT------------ISIQSSSLSG--ITWYDLDAVEIGLLTSGS 71
A GGLLFGYD+G T T + Q SG + D + L TS
Sbjct: 258 AMGGLLFGYDLGITGGVTSMEPFLVKFFPGVYKQMKDESGHESEYCKFDNELLTLFTSSL 437
Query: 72 LYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIG 130
ALI S A LGR+ + L +LVGAL+ FA N +L+IGRL+ G G+G
Sbjct: 438 YLAALIASFFASTTTRMLGRKTSMFAGGLFFLVGALLNGFAVNIEMLIIGRLLLGFGVG 614
>TC9536 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (17%)
Length = 537
Score = 67.4 bits (163), Expect = 5e-12
Identities = 32/100 (32%), Positives = 62/100 (62%)
Frame = +2
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
A+++ V +++Y + +++GP+ ++ +EIFP R+RG ++I LV + + +VT+
Sbjct: 17 AIISTVCVVVYFCFFVMAYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIVTYTLPV 196
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
+ +G +F +++ + S VF+Y VPETKG+ LE I
Sbjct: 197 MLSSMGLAGVFGLYAIVCCISWVFVYLKVPETKGMPLEVI 316
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.328 0.145 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,001,497
Number of Sequences: 28460
Number of extensions: 111896
Number of successful extensions: 772
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 740
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 749
length of query: 477
length of database: 4,897,600
effective HSP length: 94
effective length of query: 383
effective length of database: 2,222,360
effective search space: 851163880
effective search space used: 851163880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC142222.8


