

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.5 + phase: 0
(378 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
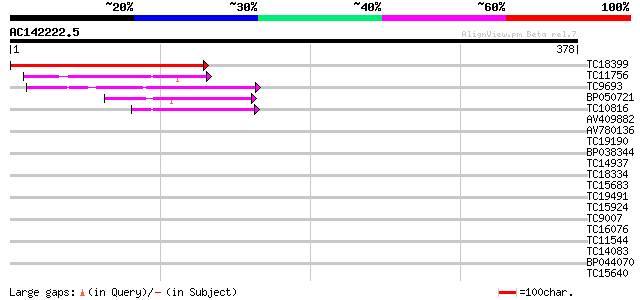
Score E
Sequences producing significant alignments: (bits) Value
TC18399 242 8e-65
TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snR... 54 4e-08
TC9693 similar to UP|COPP_MOUSE (O55029) Coatomer beta' subunit ... 50 8e-07
BP050721 45 2e-05
TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subun... 40 8e-04
AV409882 39 0.002
AV780136 38 0.003
TC19190 weakly similar to PIR|T46032|T46032 WD-40 repeat regulat... 37 0.007
BP038344 36 0.012
TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5,... 35 0.027
TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1 (AT3... 33 0.078
TC15683 weakly similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (4%) 32 0.13
TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Ar... 32 0.23
TC15924 weakly similar to GB|BAC21577.1|24060129|AP005438 G-prot... 31 0.30
TC9007 similar to UP|Q9SSS7 (Q9SSS7) F6D8.2 protein (At1g52730),... 31 0.30
TC16076 weakly similar to GB|AAG32022.1|11141605|AF277458 LEUNIG... 30 0.51
TC11544 homologue to UP|Q9SW94 (Q9SW94) G protein beta subunit, ... 30 0.51
TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor... 30 0.66
BP044070 30 0.66
TC15640 similar to GB|AAP37794.1|30725544|BT008435 At3g63460 {Ar... 30 0.86
>TC18399
Length = 542
Score = 242 bits (617), Expect = 8e-65
Identities = 113/132 (85%), Positives = 123/132 (92%)
Frame = +1
Query: 1 MEINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAA 60
MEINLGKL FDIDFHPSDNLVA GLI GD H+YRYSSD+ ++DPVRLLE+HAHTESCRAA
Sbjct: 145 MEINLGKLGFDIDFHPSDNLVAAGLISGDFHIYRYSSDSNDNDPVRLLEVHAHTESCRAA 324
Query: 61 RFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
RFINGGR LLTGSPD SILATDVETGSTI R+DNAHEAA+NRL+NLTESTVASGDD+GCI
Sbjct: 325 RFINGGRVLLTGSPDCSILATDVETGSTITRIDNAHEAAINRLINLTESTVASGDDEGCI 504
Query: 121 KVWDTRERSCCN 132
KVWD RE+SCCN
Sbjct: 505 KVWDIREQSCCN 540
>TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snRNP-specific
40 kDa protein (hPrp8-binding) {Homo sapiens;} , partial
(43%)
Length = 719
Score = 53.9 bits (128), Expect = 4e-08
Identities = 32/127 (25%), Positives = 60/127 (47%), Gaps = 2/127 (1%)
Frame = +1
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F+P+ ++A+G D ++ L+ N + + + + H + + G +
Sbjct: 316 YTMKFNPAGTVIASGSHDREIFLW-----NVHGECKNFMVLKGHKNAVLDLHWTTDGTQI 480
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST--VASGDDDGCIKVWDTRE 127
++ SPD ++ DVETG + ++ H + VN V SG DDG K+WD R+
Sbjct: 481 VSASPDKTVRVWDVETGKQVKKMVE-HLSYVNSCCPSRRGPPLVVSGSDDGTAKLWDMRQ 657
Query: 128 RSCCNSF 134
R +F
Sbjct: 658 RGSIQTF 678
>TC9693 similar to UP|COPP_MOUSE (O55029) Coatomer beta' subunit
(Beta'-coat protein) (Beta'-COP) (p102), partial (20%)
Length = 743
Score = 49.7 bits (117), Expect = 8e-07
Identities = 41/159 (25%), Positives = 72/159 (44%), Gaps = 3/159 (1%)
Frame = +1
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+D HP++ + L G + ++ Y + T + + E+ R+A+FI + ++
Sbjct: 268 VDLHPTEPWILASLYSGTVCIWNYQTQ-TMAKSFEVTELPV-----RSAKFIARKQWVVA 429
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSC 130
G+ D I + T + ++ AH + + V+ T V S DD IK+WD +
Sbjct: 430 GADDMFIRVYNYNTMDKV-KVFEAHTDYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGWI 606
Query: 131 CNSF-EAHEDYISDITF-ASDAMKLLATSGDGTLSVCNL 167
C EAH Y+ +TF D + S D T+ + NL
Sbjct: 607 CTQICEAHSHYVMQVTFNPKDTNTFASASLDRTIRIRNL 723
>BP050721
Length = 562
Score = 45.1 bits (105), Expect = 2e-05
Identities = 28/107 (26%), Positives = 49/107 (45%), Gaps = 6/107 (5%)
Frame = -1
Query: 64 NGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNL------TESTVASGDDD 117
N G +L G D ++ D +G + ++ N+ L T++ VA G +D
Sbjct: 481 NDGNCILAGCLDSTVRLLDRTSGELLQE----YKGHTNKSYKLDCCLTNTDAHVAGGSED 314
Query: 118 GCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
G I WD + +SF AH ++ +++ +L+ S DGT+ V
Sbjct: 313 GFIYFWDLVDAYVVSSFRAHTSVVTSVSYHPKENCMLSASVDGTIRV 173
>TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subunit-like
protein, partial (20%)
Length = 979
Score = 39.7 bits (91), Expect = 8e-04
Identities = 24/86 (27%), Positives = 42/86 (47%), Gaps = 1/86 (1%)
Frame = +1
Query: 82 DVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDY 140
D G+ I + D H+ V + + ++ SG DD IKVW+ + C + H DY
Sbjct: 349 DYRMGTLIDKFDE-HDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDY 525
Query: 141 ISDITFASDAMKLLATSGDGTLSVCN 166
I + F ++ +++ S D T+ + N
Sbjct: 526 IRTVQFHHESPWIVSASDDQTIRIWN 603
Score = 35.4 bits (80), Expect = 0.016
Identities = 28/156 (17%), Positives = 57/156 (35%)
Frame = +1
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ FHP + L G + L+ Y + + H R F + ++
Sbjct: 283 LSFHPKRPWILASLHSGVIQLWDYRMGTL------IDKFDEHDGPVRGVHFHHSQPLFVS 444
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCC 131
G D+ I + + + L + + + S DD I++W+ + R+C
Sbjct: 445 GGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCI 624
Query: 132 NSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNL 167
+ H Y+ F +++ S D T+ V ++
Sbjct: 625 SVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDI 732
>AV409882
Length = 422
Score = 38.5 bits (88), Expect = 0.002
Identities = 32/128 (25%), Positives = 57/128 (44%), Gaps = 2/128 (1%)
Frame = +3
Query: 21 VATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILA 80
+ L G + ++ Y S T + + E+ R+A+F+ + ++ G+ D I
Sbjct: 15 ILASLYSGTVCIWNYQSQ-TMAKSFEVTELPV-----RSAKFVARKQWVVAGADDMFIRV 176
Query: 81 TDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSCCNS-FEAHE 138
+ T + ++ AH + + V+ T V S DD IK+WD + C FE H
Sbjct: 177 YNYNTMDKV-KVFEAHTDYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGWICTQIFEGHS 353
Query: 139 DYISDITF 146
Y+ +TF
Sbjct: 354 HYVMQVTF 377
Score = 28.9 bits (63), Expect = 1.5
Identities = 16/49 (32%), Positives = 24/49 (48%)
Frame = +3
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGD 159
V +G DD I+V++ FEAH DYI + +L++S D
Sbjct: 141 VVAGADDMFIRVYNYNTMDKVKVFEAHTDYIRCVAVHPTLPYVLSSSDD 287
>AV780136
Length = 558
Score = 37.7 bits (86), Expect = 0.003
Identities = 20/77 (25%), Positives = 35/77 (44%)
Frame = -1
Query: 65 GGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWD 124
GG L++G + I D +GS +L + L++ T SG D I++WD
Sbjct: 540 GGTVLVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDSTGRYCLSGSSDSMIRLWD 361
Query: 125 TRERSCCNSFEAHEDYI 141
+ C +++ H D +
Sbjct: 360 IGQPRCLHTYAVHTDSV 310
Score = 27.7 bits (60), Expect = 3.3
Identities = 15/59 (25%), Positives = 25/59 (41%)
Frame = -1
Query: 109 STVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNL 167
+ + SG + I+VWD R S H D I + S L+ S D + + ++
Sbjct: 534 TVLVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDSTGRYCLSGSSDSMIRLWDI 358
>TC19190 weakly similar to PIR|T46032|T46032 WD-40 repeat regulatory protein
tup1 homolog - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (18%)
Length = 511
Score = 36.6 bits (83), Expect = 0.007
Identities = 14/54 (25%), Positives = 27/54 (49%)
Frame = +3
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
+ SG +D C+ +WD ++++ E H D + +T K+ + DG +V
Sbjct: 24 IVSGSEDRCVYLWDLQQKNMIQKLEGHTDTVISVTCHPTENKIASAGLDGDRTV 185
>BP038344
Length = 525
Score = 35.8 bits (81), Expect = 0.012
Identities = 21/71 (29%), Positives = 37/71 (51%), Gaps = 1/71 (1%)
Frame = +2
Query: 95 AHEAAVNRLVNLTESTV-ASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKL 153
AH+AAV + + T+ AS D + +W + S + H + ISD+ ++SD+ +
Sbjct: 92 AHDAAVACVKFSNDGTLLASASLDKTLIIWSSATLSLLHRLTGHSEGISDLAWSSDSHYI 271
Query: 154 LATSGDGTLSV 164
+ S D TL +
Sbjct: 272 CSASDDRTLRI 304
>TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5, TAC
clone:K8K14 (AT5g67320/K8K14_4), partial (19%)
Length = 1089
Score = 34.7 bits (78), Expect = 0.027
Identities = 19/85 (22%), Positives = 37/85 (43%)
Frame = +1
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+AS D +K+WD S H D + + F+ + L + S D ++ + +L+
Sbjct: 55 LASASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSLKEG 234
Query: 171 KVQAQSEFSEDELLSVVLMKNGRKV 195
K+ ++ + V K G K+
Sbjct: 235 KI-VKTYTGSGGIFEVCWNKEGDKI 306
Score = 32.0 bits (71), Expect = 0.17
Identities = 22/100 (22%), Positives = 42/100 (42%)
Frame = +1
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRER 128
L + S D ++ DVE G I L+ + + + +ASG D I +W +E
Sbjct: 55 LASASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSLKEG 234
Query: 129 SCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLR 168
++ I ++ + + K+ A + T+ V + R
Sbjct: 235 KIVKTYTG-SGGIFEVCWNKEGDKIAACFANNTVCVLDFR 351
>TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1
(AT3g18860/MCB22_3), partial (21%)
Length = 595
Score = 33.1 bits (74), Expect = 0.078
Identities = 33/130 (25%), Positives = 55/130 (41%), Gaps = 7/130 (5%)
Frame = +2
Query: 49 EIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIA-RLDNAHEAAVNRLV--- 104
E+H H + R + G + T S D ++ E ++ ++ H + V LV
Sbjct: 134 ELHGHEDDVRGI-CVCGNDGIATSSRDKTVRFWSPENRKFVSSKVLVGHSSFVGPLVWIP 310
Query: 105 ---NLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGT 161
L + VASG D + VWD ++ + H+ ++ I F D +++ S D
Sbjct: 311 PNPELPQGGVASGGMDTLVLVWDLSTGEKAHTLKGHQLQVTSIAF--DDGDVVSASVD-- 478
Query: 162 LSVCNLRRNK 171
C LRR K
Sbjct: 479 ---CTLRRWK 499
>TC15683 weakly similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (4%)
Length = 957
Score = 32.3 bits (72), Expect = 0.13
Identities = 31/155 (20%), Positives = 60/155 (38%), Gaps = 11/155 (7%)
Frame = +2
Query: 40 TNSDPVRLLEIHAHTESCRAARFIN----------GGRALLTGSPDFSILATDVETGSTI 89
TNS VRL +I A + F G + G+ + + D+ TG +
Sbjct: 41 TNSHQVRLYDISAQRRPVLSFDFRETPIKALAEDIDGYTVYIGNGSGDMASVDIRTGKML 220
Query: 90 ARLDNAHEAAVNRLVNLTE-STVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFAS 148
++ +V E +AS D +++WDT+ R ++ + + + +S
Sbjct: 221 GGFTGKCSGSIRSIVRHPELPVIASCGLDSYLRIWDTKTRQLLSAVFLKQPVLHALFDSS 400
Query: 149 DAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDEL 183
++ ++ DG S N++ A+ E L
Sbjct: 401 FIVEEPSSGADGLPSKAQ-NLNEIAAEEEVEPSPL 502
>TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;}, partial (39%)
Length = 527
Score = 31.6 bits (70), Expect = 0.23
Identities = 17/80 (21%), Positives = 33/80 (41%)
Frame = +1
Query: 67 RALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTR 126
R L T S D ++ D E + + + + V+ +A+G D +++WD
Sbjct: 58 RLLFTASDDGNVHMYDAEGKALVGTMSGHASWVLCVDVSPDGGAIATGSSDRTVRLWDLA 237
Query: 127 ERSCCNSFEAHEDYISDITF 146
R+ + H D + + F
Sbjct: 238 MRASVQTMSNHTDQVWGVAF 297
>TC15924 weakly similar to GB|BAC21577.1|24060129|AP005438 G-protein beta
family-like protein {Oryza sativa (japonica
cultivar-group);}, partial (11%)
Length = 1262
Score = 31.2 bits (69), Expect = 0.30
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Frame = +1
Query: 113 SGDDDGCIKVWDTRERSCCN--SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
SG DG IKVWD R S + H+ ++ + + LL+ S D T+ V + +
Sbjct: 124 SGYSDGSIKVWDIRGHSASLVWDIKEHKKSVTCFSVHEPSDSLLSGSTDKTIKVWKMIQR 303
Query: 171 KVQ 173
K++
Sbjct: 304 KLE 312
>TC9007 similar to UP|Q9SSS7 (Q9SSS7) F6D8.2 protein (At1g52730), partial
(23%)
Length = 647
Score = 31.2 bits (69), Expect = 0.30
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Frame = +3
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWD 124
G + G D I D TG+ IA + H V+ + + + ASG +DG I++W
Sbjct: 57 GNKFIAGGEDMWIHVFDFHTGNEIA-CNKGHHGPVHCVRFSPGGESYASGSEDGTIRIWQ 233
Query: 125 T 125
T
Sbjct: 234 T 236
>TC16076 weakly similar to GB|AAG32022.1|11141605|AF277458 LEUNIG
{Arabidopsis thaliana;} , partial (24%)
Length = 1048
Score = 30.4 bits (67), Expect = 0.51
Identities = 34/193 (17%), Positives = 82/193 (41%), Gaps = 2/193 (1%)
Frame = +3
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+AS DD + +W+ ++ E H+ ISD+ F ++ +L S D ++ + + N
Sbjct: 39 LASAGDDKKVVLWNMDTLQTESTPEQHKSVISDVRFRPNSSQLATASIDKSVRLWD-AAN 215
Query: 171 KVQAQSEFS--EDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSID 228
E++ ++S+ + C + + Y W R S+ +
Sbjct: 216 PTYCVQEYNGHSSAIMSLDFHPKKTDIFCFCDSANEIRY-WNITSSSCTRVSKGGSSQVR 392
Query: 229 TMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLG 288
++ + +++ + + ++++ + +R I + H E PV + + + FL
Sbjct: 393 FQPRIGQ--VLSAASD*VVSIFDVESDRQIYTLQGHPE-PVNS------ICWDVNGDFLA 545
Query: 289 SIAHDQMLKLWDL 301
S++ ++K+W L
Sbjct: 546 SVS-PNLVKIWSL 581
>TC11544 homologue to UP|Q9SW94 (Q9SW94) G protein beta subunit, partial
(50%)
Length = 774
Score = 30.4 bits (67), Expect = 0.51
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 6/67 (8%)
Frame = +2
Query: 69 LLTGSPDFSILATDVETGSTIA----RLDNAHEAAVNRL-VNLTESTV-ASGDDDGCIKV 122
L+TGS D + + D+ TG + + H A V + +N + S + SG DG ++
Sbjct: 572 LITGSGDQTCVLWDITTGLRTSVFGGEFQSGHTADVLSISINGSNSRMFVSGSCDGTARL 751
Query: 123 WDTRERS 129
WDTR S
Sbjct: 752 WDTRVAS 772
>TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor, partial
(92%)
Length = 1693
Score = 30.0 bits (66), Expect = 0.66
Identities = 21/101 (20%), Positives = 38/101 (36%)
Frame = +1
Query: 278 VAFSHDRKFLGSIAHDQMLKLWDLDNILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNT 337
V + D ++ +A + K D + +R E ++ DSD EDE D D +
Sbjct: 1108 VLITDDPEYAKQVAEETWGKQKDAEKAAFEEAEKKREEEESKEDPADSDAEDEEDTDEAS 1287
Query: 338 SKFTKGNKRKNASKGHAAGDSNNFFADL*EIMAATFSTTSS 378
+ A + A + + +L* + A + S
Sbjct: 1288 HDSDDAESKTEAGEDSADANEEDVHDEL*RV*KADTGVSES 1410
>BP044070
Length = 289
Score = 30.0 bits (66), Expect = 0.66
Identities = 20/81 (24%), Positives = 41/81 (49%), Gaps = 6/81 (7%)
Frame = +2
Query: 88 TIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVW-----DTRERSCCNSFEAHEDYI 141
T+++ H+ A++ + + +AS D ++ + D+ S +E H+ +
Sbjct: 44 TLSQTLTGHKRAISAVKFSSNGRLLASSSADKTLRTYGFTTSDSVTLSPMQLYEGHDXGV 223
Query: 142 SDITFASDAMKLLATSGDGTL 162
SD+ F+SD+ L++ S D TL
Sbjct: 224 SDLAFSSDSRFLVSASDDKTL 286
>TC15640 similar to GB|AAP37794.1|30725544|BT008435 At3g63460 {Arabidopsis
thaliana;}, partial (26%)
Length = 1022
Score = 29.6 bits (65), Expect = 0.86
Identities = 23/74 (31%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Frame = +2
Query: 106 LTESTVASGDDDGCIKVWDTRERSCCNSF-------EAHEDYISDITFASDAMKLLA-TS 157
+ + +ASG +DG I +WD S F A + IS +++ S +LA TS
Sbjct: 527 IAPNLLASGAEDGEICIWDLANPSEPTHFPPLKGSGSASQGEISFLSWNSKVQHILASTS 706
Query: 158 GDGTLSVCNLRRNK 171
+GT V +L++ K
Sbjct: 707 YNGTTVVWDLKKQK 748
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,471,565
Number of Sequences: 28460
Number of extensions: 64195
Number of successful extensions: 407
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 385
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 406
length of query: 378
length of database: 4,897,600
effective HSP length: 92
effective length of query: 286
effective length of database: 2,279,280
effective search space: 651874080
effective search space used: 651874080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC142222.5


