

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142095.2 + phase: 0
(458 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
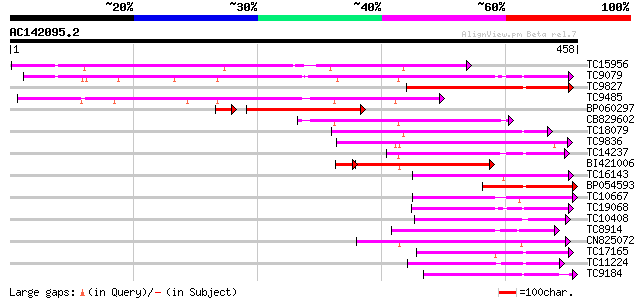
Score E
Sequences producing significant alignments: (bits) Value
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 207 2e-54
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 172 1e-43
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 157 3e-39
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 143 5e-35
BP060297 125 2e-32
CB829602 115 2e-26
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 114 3e-26
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 114 3e-26
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 107 4e-24
BI421006 98 4e-23
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 98 2e-21
BP054593 97 7e-21
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 93 1e-19
TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 90 9e-19
TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransfe... 90 9e-19
TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl tr... 89 1e-18
CN825072 87 6e-18
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 87 7e-18
TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase... 87 7e-18
TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone glu... 82 1e-16
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 207 bits (528), Expect = 2e-54
Identities = 139/398 (34%), Positives = 199/398 (49%), Gaps = 26/398 (6%)
Frame = +2
Query: 2 ENKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPN-- 59
E K S +H L++ FPA GH NP+L +K L + G VTL TT + K++ N
Sbjct: 8 EKKMGSESPLHILLVSFPAQGHINPLLRLAKCLAAK-GSSVTLATTQTAGKSMRTATNIT 184
Query: 60 ---------NSITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDH 110
S+ E DG + + LYL + VG Q + +I N
Sbjct: 185 DKAPTPISDGSLKFEFFDDGLGPDDDSIRGNLSLYLQQLELVGRQQILKMIKEHADSNRQ 364
Query: 111 VDCLIYDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPAL 170
+ C+I + F+PW +DVA+E GI A Q+ + + YY + P E + L
Sbjct: 365 ISCIINNPFLPWVVDVAEEQGIPSALLWIQSSAVFTAYYSYFHNLVTFPSKEDPLADVQL 544
Query: 171 PQ----LQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMK 226
P L+ ++P F P + + QF N++K IL +++ ELE + D+ K
Sbjct: 545 PSTSIVLKHCEIPDFLHPSCTYPFLGTLILEQFKNLNKTFCILVDTYEELEHDFIDYLSK 724
Query: 227 IWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQ----LKSDESIEWLNNKPKRSAVYVSFGS 282
+ R VGP K + S A +KSD+ IEWLN+KP+ S VYVSFGS
Sbjct: 725 TFL-IRPVGPLF---------KSSQAKSAAIRGDFMKSDDCIEWLNSKPQSSVVYVSFGS 874
Query: 283 MASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETK------LPKDF-EKKSENGLVVAWCP 335
+ L +EQ++E+A LK FLWV+K E LP F E+ E G VV W P
Sbjct: 875 IVYLPQEQVDEIAFGLKKSQVSFLWVMKPPPEVTGLQPHVLPYGFLEETGERGRVVQWSP 1054
Query: 336 QLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLY 373
Q EVLAH ++ CF+THCGWNS++EAL+ GVP++ P +
Sbjct: 1055QEEVLAHPSVSCFLTHCGWNSSMEALTSGVPVLTFPAW 1168
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 172 bits (435), Expect = 1e-43
Identities = 129/476 (27%), Positives = 231/476 (48%), Gaps = 32/476 (6%)
Frame = +1
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKL--PNN-----SITI 64
H ++ P P GH NP+ + +KLL + G +T V T N+K + K PN
Sbjct: 19 HAVLTPSPLQGHINPLFQLAKLLHLR-GFHITFVHTEYNHKRLIKSRGPNALDGLPDFVF 195
Query: 65 ETISDGFDKGGVAEAKDFKLYL----NKFWQVGPQSLAHLINNLNARN-DHVDCLIYDSF 119
ETI DG + G ++D NK LA L + +A V CL+ D
Sbjct: 196 ETIPDGLEDGDGEVSQDLYSLCDSIRNKCHLPFQDLLAKLHRSASAGLVPPVTCLVSDLL 375
Query: 120 MPWCLDVAKEFG--IVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEIT----------- 166
M + + A++ I+ + + M+ +++ L K P ++
Sbjct: 376 MTFTIQAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNGYLDTKVDW 555
Query: 167 LPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMK 226
+P + + +D+P F T + + L+ V +A I+ N+F ELE++V
Sbjct: 556 IPGMQNFRLKDLPDFIRTTDPNDIMLEFMVEVADRAKRASAIVFNTFNELERDVLSALST 735
Query: 227 IWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDES--IEWLNNKPKRSAVYVSFGSMA 284
+ + +GP P +FL++ ++ + L +++ ++WL +K S VYV+FGS+
Sbjct: 736 MLPSLYPIGP-FP-SFLNQAPQNQLASLGSNLWKEDTKCLQWLESKEPGSVVYVNFGSIT 909
Query: 285 SLNEEQIEEVAHCLKDCGSYFLWVVKTS-----EETKLPKDFEKKSENGLVVAWCPQLEV 339
++ EQ+ E A L + FLW+++ PK ++ S GL+ +WCPQ +V
Sbjct: 910 VMSPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPKFVDETSNRGLIASWCPQEKV 1089
Query: 340 LAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQ 399
L H ++G F+THCGWNST+E++ GVP++ P +DQ + +++ + W +GI VD
Sbjct: 1090LNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDIGIE--VDTN- 1260
Query: 400 IVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
V+++ +++ I E+M + +KGK++ M+ K A G S+ N+ + + +
Sbjct: 1261-VKREEVENLIHELM-VGDKGKKMRQRTMELKKKAEEDTRPGGCSYMNLDKLIKEV 1422
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 157 bits (398), Expect = 3e-39
Identities = 69/135 (51%), Positives = 103/135 (76%)
Frame = +2
Query: 321 FEKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDA 380
F + +E GL+V WCPQL VL H+A+GCF+THCGWNSTLE++S+GVP++A+PL++D +
Sbjct: 5 FVETTEKGLIVTWCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPVTNG 184
Query: 381 KFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGK 440
K +VD+WK G+R + DEK+IVR++ +K+CI EI+ +EK EI +N ++W LA ++ +
Sbjct: 185 KLIVDVWKTGVRAVADEKEIVRRETIKNCIKEIIE-TEKINEIKSNAIKWMNLAKDSLAE 361
Query: 441 DGSSHKNMIEFVNSL 455
G S KN+ EFV +L
Sbjct: 362 GGRSDKNIAEFVAAL 406
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 143 bits (361), Expect = 5e-35
Identities = 114/381 (29%), Positives = 177/381 (45%), Gaps = 36/381 (9%)
Frame = +1
Query: 7 STKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPK---------L 57
+ K H + +P+PA GH NPML+ +KLL + G VT V T N+K + K L
Sbjct: 76 AAKKPHVVCIPYPAQGHINPMLKLAKLLHFKGGFHVTFVNTEYNHKRLLKSRGPDSLNGL 255
Query: 58 PNNSITIETISDGFDKGGVAEAKDFK---LYLNKFWQVGPQSLAHLINNLNARNDHVDCL 114
P S ETI DG + V +D + K + L +N++++ V C+
Sbjct: 256 P--SFRFETIPDGLPETDVDVTQDIPSLCISTRKTCLPHFKKLLSKLNDVSSDVPPVTCI 429
Query: 115 IYDSFMPWCLDVAKEFGIVGASFLTQNL--VMNSIYYHVHLGK-LKPPFVEQEIT----- 166
+ D M + LD A E I F T + M + Y + K + P +IT
Sbjct: 430 VSDGCMSFTLDAAIELNIPEVLFWTTSACGFMGYVQYRELIEKGIIPLKDSSDITNGYLE 609
Query: 167 -----LPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVA 221
LP + ++ +D+PSF T + + LD + KA I+ N+F LE +V
Sbjct: 610 TTIEWLPGMKNIRLKDLPSFLRTTDPNDKMLDFLTGECQRALKASAIILNTFDALEHDVL 789
Query: 222 DWTMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSD------ESIEWLNNKPKRSA 275
+ I ++GP L +KD D ++ L S+ E ++WL+ K S
Sbjct: 790 EAFSSILPPVYSIGP------LHLLIKDVTDKNLNSLGSNLWKEDSECLKWLDTKEPNSV 951
Query: 276 VYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVK----TSEETKLPKDF-EKKSENGLV 330
VYV+FGS+ + EQ+ E A L + FLWV++ + LP++F ++ G +
Sbjct: 952 VYVNFGSITVMTSEQMVEFAWGLANSNKTFLWVIRPDLVAGKHAVLPEEFVAATNDRGRL 1131
Query: 331 VAWCPQLEVLAHEAIGCFVTH 351
+W PQ +VL H AIG F+TH
Sbjct: 1132SSWTPQEDVLTHPAIGGFLTH 1194
>BP060297
Length = 364
Score = 125 bits (314), Expect(2) = 2e-32
Identities = 57/96 (59%), Positives = 73/96 (75%)
Frame = +2
Query: 192 LDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRVKDDE 251
L++ VAQFSNI KAD ILCNSF+EL+KEV DWTMK FRT+GP +P FL K +KDDE
Sbjct: 77 LELVVAQFSNIDKADXILCNSFYELDKEVTDWTMKTXPKFRTIGPSIPSMFLGKEIKDDE 256
Query: 252 DHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLN 287
D+ +A+ S++ + WLN++PK S VYV FGS +LN
Sbjct: 257 DYGVAKFTSEDYMAWLNDRPKGSVVYVCFGSYGALN 364
Score = 30.8 bits (68), Expect(2) = 2e-32
Identities = 12/17 (70%), Positives = 15/17 (87%)
Frame = +1
Query: 167 LPALPQLQPRDMPSFYF 183
LP LP+LQ +DMPSF+F
Sbjct: 1 LPGLPKLQHQDMPSFFF 51
>CB829602
Length = 542
Score = 115 bits (288), Expect = 2e-26
Identities = 56/186 (30%), Positives = 109/186 (58%), Gaps = 11/186 (5%)
Frame = +1
Query: 233 TVGPCLPYTFLDKRVKDDEDHSIAQLKSD------ESIEWLNNKPKRSAVYVSFGSMASL 286
T+GP LD + ++ I +K + E ++WL+++ S +YV+FGS+ ++
Sbjct: 4 TIGP------LDLLLNQVTENRIESIKCNLWKEEPECLKWLDSQEPSSVLYVNFGSVINM 165
Query: 287 NEEQIEEVAHCLKDCGSYFLWVVKTS-----EETKLPKDFEKKSENGLVVAWCPQLEVLA 341
+Q+ E+A + + F+WV++ LP+ + + G++++WCPQ +VL
Sbjct: 166 TPQQLVELAWGIANSKKKFIWVIRPDLVEGEASIVLPEIVAETKDRGIMLSWCPQEQVLK 345
Query: 342 HEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIV 401
H A+G F+THCGWNST+E++S GVP++ P ++DQ ++++++ W G+ D+ V
Sbjct: 346 HSALGGFLTHCGWNSTIESISSGVPLICSPFFNDQFVNSRYICSEWDFGMEMNSDD---V 516
Query: 402 RKDPLK 407
++D ++
Sbjct: 517 KRDEVE 534
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 114 bits (286), Expect = 3e-26
Identities = 72/196 (36%), Positives = 108/196 (54%), Gaps = 18/196 (9%)
Frame = +3
Query: 261 DESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETK---- 316
DE + WL+ +P S +YVSFGS ++ + Q+ E+A L+ F+WVV+ E
Sbjct: 54 DEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQHRFVWVVRPPNEADASAT 233
Query: 317 ------------LPKDFEKKS-ENGLVVA-WCPQLEVLAHEAIGCFVTHCGWNSTLEALS 362
LP+ F ++ E G+VV W PQ E+L H A G FVT CGWNS LE++
Sbjct: 234 FFGFGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEILGHPATGGFVTPCGWNSVLESVL 413
Query: 363 IGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKE 422
GVP+VA PLYS+Q ++A L + V +R V E +V +D + D + +M +SE+G
Sbjct: 414 NGVPMVAWPLYSEQKMNAYMLSEELGVAVRVKVAEGGVVCRDQVVDMVRRVM-VSEEGMG 590
Query: 423 IMNNVMQWKTLATRAV 438
+ + V + K A+
Sbjct: 591 MKDRVRELKLSGDEAL 638
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 114 bits (286), Expect = 3e-26
Identities = 73/206 (35%), Positives = 118/206 (56%), Gaps = 16/206 (7%)
Frame = +1
Query: 265 EWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVK-------TSE---E 314
+WL+ KP S ++VSFGSM +++ Q+ ++A L G F+WVV+ SE E
Sbjct: 385 KWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEFRAE 564
Query: 315 TKLPKDF-EKKSENGLVVA-WCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPL 372
LP+ F + ++ GLVV W PQ+E+L+H A+ F++HCGWNS LE+LS GVPI+ P+
Sbjct: 565 EWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILGWPM 744
Query: 373 YSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKT 432
++Q + K L + V + ++ VR + L + I +M+ +E G +I N +
Sbjct: 745 AAEQFFNCKMLEEELGVCVEVARGKRCEVRHEDLVEKIELVMNEAESGVKIRKNAGNIRE 924
Query: 433 LATRAV----GKDGSSHKNMIEFVNS 454
+ AV G GSS + + EF+++
Sbjct: 925 MIRDAVRDEDGYKGSSVRAIDEFLSA 1002
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 107 bits (267), Expect = 4e-24
Identities = 51/154 (33%), Positives = 91/154 (58%), Gaps = 6/154 (3%)
Frame = +2
Query: 305 FLWVVKTS-----EETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTL 358
FLW+++ + +P++F ++ G + +WC Q EVL+H +IG F+THCGWNST+
Sbjct: 218 FLWILRPDVVMGEDSINVPQEFLDEIKGRGYIASWCFQEEVLSHPSIGAFLTHCGWNSTI 397
Query: 359 EALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSE 418
E +S G+P++ P +S+Q ++++ W++G+ D V++D + + E+M E
Sbjct: 398 EGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHD----VKRDEITTLVNEMMK-GE 562
Query: 419 KGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 452
KGKE+ ++WK A A G GSS+ + + +
Sbjct: 563 KGKEMRKKGLEWKKKAIEATGLGGSSYNDFHKLI 664
>BI421006
Length = 519
Score = 98.2 bits (243), Expect(2) = 4e-23
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 13/125 (10%)
Frame = +2
Query: 280 FGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSE-----------ETKLPKDFEKKSEN- 327
F ++ EEQIE++A+ L+ F+WV++ ++ E LPK FEK+ E
Sbjct: 101 FWFFSTFIEEQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGFEKRVEGM 280
Query: 328 GLVVA-WCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDI 386
GLVV W PQLE+L+H + G F++HCGWNS +E++S+GVPI A P++SDQ + + ++
Sbjct: 281 GLVVRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEV 460
Query: 387 WKVGI 391
KV +
Sbjct: 461 LKVAL 475
Score = 26.6 bits (57), Expect(2) = 4e-23
Identities = 9/18 (50%), Positives = 15/18 (83%)
Frame = +3
Query: 264 IEWLNNKPKRSAVYVSFG 281
+EWL+ + ++S +YVSFG
Sbjct: 54 MEWLDRQEQKSVLYVSFG 107
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 98.2 bits (243), Expect = 2e-21
Identities = 44/140 (31%), Positives = 83/140 (58%), Gaps = 10/140 (7%)
Frame = +1
Query: 326 ENGLVV-AWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLV 384
E G+++ W PQ+ +L H A+G FVTHCGWNST+EA+S GVP++ P+ +Q + K +
Sbjct: 4 EKGMIIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLIT 183
Query: 385 DIWKVGIRPLVDE---------KQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLAT 435
++ +G+ +E +++V ++ ++ + +M ++G++I ++ A
Sbjct: 184 EVRGIGVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGEKAR 363
Query: 436 RAVGKDGSSHKNMIEFVNSL 455
+AV + GSSHKN+ ++ L
Sbjct: 364 QAVEEGGSSHKNLTALIDDL 423
>BP054593
Length = 507
Score = 96.7 bits (239), Expect = 7e-21
Identities = 47/76 (61%), Positives = 58/76 (75%)
Frame = -3
Query: 383 LVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDG 442
+VD+WK GIR +DEKQ+VR+D LK CI E+M SE GKEI +N MQWK LA AV K G
Sbjct: 502 IVDVWKSGIRAPLDEKQVVRRDALKHCIWELME-SENGKEIKSNAMQWKNLAAAAVSKGG 326
Query: 443 SSHKNMIEFVNSLFQV 458
SSHK++ EFV+SLF +
Sbjct: 325 SSHKHITEFVDSLFHL 278
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 92.8 bits (229), Expect = 1e-19
Identities = 47/136 (34%), Positives = 78/136 (56%), Gaps = 3/136 (2%)
Frame = +1
Query: 326 ENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVD 385
E G V W PQ +VL H AIG F THCGWNSTLE++ GVP++ P + DQ ++AK++ D
Sbjct: 1 EFGTSVKWAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSD 180
Query: 386 IWKVGIRPLVDEKQIVRKDPLKDCI---CEIMSMSEKGKEIMNNVMQWKTLATRAVGKDG 442
+WKVG++ ++ P + I ++ + ++ EI N+++ K A + + G
Sbjct: 181 VWKVGVQ--------LQNKPGRGEIEKTISLLMLGDEAYEIRGNILKLKEKANVCLSEGG 336
Query: 443 SSHKNMIEFVNSLFQV 458
SS+ + V+ + +
Sbjct: 337 SSYCFLDRLVSEILSL 384
>TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (26%)
Length = 565
Score = 89.7 bits (221), Expect = 9e-19
Identities = 44/131 (33%), Positives = 78/131 (58%)
Frame = +1
Query: 325 SENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLV 384
S GL+ +WC Q VL H +IG F+THCGWNST+E++S GVP++ P ++DQ + + +
Sbjct: 43 SSRGLIASWCSQEHVLNHPSIGGFLTHCGWNSTIESISAGVPMLCWPFFADQLTNCRSIC 222
Query: 385 DIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSS 444
+ W +G++ +D +++ ++ I E+M + EKGK++ M+ K A G S
Sbjct: 223 NEWDIGMQ--IDTNG--KREEVEKLINELM-VGEKGKKMRQKTMELKKRAEEDTRPGGCS 387
Query: 445 HKNMIEFVNSL 455
+ N+ + + +
Sbjct: 388 YMNLDQVIKEV 420
>TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (19%)
Length = 571
Score = 89.7 bits (221), Expect = 9e-19
Identities = 40/126 (31%), Positives = 72/126 (56%)
Frame = +1
Query: 328 GLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIW 387
G VV W PQ ++L H AI CF++HCGWNST+E + VP + P +DQ ++ F+ D+W
Sbjct: 16 GKVVKWGPQKKILNHPAIACFISHCGWNSTIEGVHASVPFLCWPFSTDQFLNKSFICDVW 195
Query: 388 KVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKN 447
K+G+ DE I+ K ++ + +++ +++ ++ K + + + G S N
Sbjct: 196 KIGLGLDKDENGIIPKGEIRTKVEQVI----VNEDLKARSLKLKEITINNLAEGGQSSNN 363
Query: 448 MIEFVN 453
+ +F+N
Sbjct: 364 LKKFIN 381
>TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl transferase,
partial (32%)
Length = 627
Score = 89.4 bits (220), Expect = 1e-18
Identities = 44/137 (32%), Positives = 78/137 (56%), Gaps = 1/137 (0%)
Frame = +2
Query: 309 VKTSEETKLPKDFEKKSEN-GLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPI 367
+K ++ LP F ++++N G +V W PQ ++L HE++G FVTHCG NS E++S GVP+
Sbjct: 5 MKDHPKSLLPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVPM 184
Query: 368 VAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNV 427
+ P + D G+ + + DIW++G++ E + +D L + ++ + E+GK +
Sbjct: 185 ICRPFFGDHGMAGRIIEDIWEIGVK---IEGGVFTQDGLVKSL-NLILVQEEGKIMREKA 352
Query: 428 MQWKTLATRAVGKDGSS 444
+ K A G G +
Sbjct: 353 QKVKKTVLDAAGPQGKA 403
>CN825072
Length = 712
Score = 87.0 bits (214), Expect = 6e-18
Identities = 55/195 (28%), Positives = 93/195 (47%), Gaps = 22/195 (11%)
Frame = -3
Query: 281 GSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSE----------------ETKLPKDF-EK 323
GS S + QI E+A +++ G F+W ++ + LP+ F ++
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDR 531
Query: 324 KSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFL 383
+ G V+ W PQ VL H A G FV+HCGWNSTLE++ GVPI PLY++Q +A L
Sbjct: 530 TTGIGRVIGWAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFVL 351
Query: 384 VDIWKVGIRPLVDEKQIVRKDPLKDCICE-----IMSMSEKGKEIMNNVMQWKTLATRAV 438
V K+ + +D + + P + I S+ +K E+ V + + + +
Sbjct: 350 VRELKIAVEISLDYRVEINGGPNYLLTADKIEGGIRSVLDKDGEVRKRVKEMSEKSRKTL 171
Query: 439 GKDGSSHKNMIEFVN 453
+ G S+ + ++
Sbjct: 170 LEGGCSYSYLDRLID 126
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 86.7 bits (213), Expect = 7e-18
Identities = 47/134 (35%), Positives = 76/134 (56%), Gaps = 7/134 (5%)
Frame = +1
Query: 329 LVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWK 388
++ W PQ+ +L HEAIG FVTHCGWNSTLE ++ GVP+V P+ ++Q + K + ++ K
Sbjct: 1 IIRGWAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLK 180
Query: 389 VGI-------RPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKD 441
+G+ +V E V+ D ++ + IM E+ +E+ N V A AV +
Sbjct: 181 IGVPVGAKKWGRMVLEGDGVKSDAVEKGVKRIME-GEEAEEMRNRVKVLSQQAQWAVEEG 357
Query: 442 GSSHKNMIEFVNSL 455
GSS+ ++ + L
Sbjct: 358 GSSYSDLNALIEEL 399
>TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (19%)
Length = 596
Score = 86.7 bits (213), Expect = 7e-18
Identities = 45/127 (35%), Positives = 73/127 (57%)
Frame = +3
Query: 322 EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAK 381
E+ G VV+W PQ+E+L H ++G F+TH GWNS LE + GVP++ P + DQ I+
Sbjct: 15 ERTKSQGKVVSWAPQMEILKHASVGVFLTHGGWNSVLECIVGGVPMIGRPFFGDQRINIW 194
Query: 382 FLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKD 441
L + VG+ + ++ K+ + + M+ E+GK + + + K +A +AV D
Sbjct: 195 MLAKLLGVGVGL---QNGVLAKETILKTLKSTMT-GEEGKVMRQKMAELKEMAWKAVEPD 362
Query: 442 GSSHKNM 448
GSS KN+
Sbjct: 363 GSSTKNL 383
>TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (26%)
Length = 643
Score = 82.4 bits (202), Expect = 1e-16
Identities = 45/124 (36%), Positives = 74/124 (59%)
Frame = +3
Query: 335 PQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPL 394
PQ +VL+H +IG F++HCGWNS LE+++ GVP+VA PLY++Q ++A + + KV +RP
Sbjct: 6 PQAKVLSHASIGGFLSHCGWNSVLESVANGVPLVAWPLYAEQRMNAVLVTEDVKVALRPK 185
Query: 395 VDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNS 454
V E +V + + +M E+GK++ + K A + ++GSS N
Sbjct: 186 VGENGLVELVEIARVVRTLM-QGEEGKKLRCKMKDLKEAAATTLKENGSS-------TNQ 341
Query: 455 LFQV 458
+FQ+
Sbjct: 342 IFQL 353
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,817,221
Number of Sequences: 28460
Number of extensions: 127269
Number of successful extensions: 726
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 698
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 701
length of query: 458
length of database: 4,897,600
effective HSP length: 93
effective length of query: 365
effective length of database: 2,250,820
effective search space: 821549300
effective search space used: 821549300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC142095.2


