

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.7 - phase: 0
(406 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
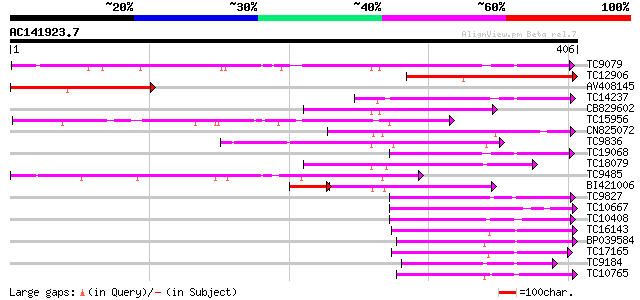
Score E
Sequences producing significant alignments: (bits) Value
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 172 9e-44
TC12906 similar to PIR|T00584|T00584 indole-3-acetate beta-gluco... 162 9e-41
AV408145 115 2e-26
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 110 3e-25
CB829602 110 3e-25
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 109 7e-25
CN825072 106 8e-24
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 105 1e-23
TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 103 7e-23
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 102 2e-22
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 99 1e-21
BI421006 89 2e-21
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 97 6e-21
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 92 2e-19
TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransfe... 91 3e-19
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 91 5e-19
BP039584 90 6e-19
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 89 1e-18
TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone glu... 81 4e-16
TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid gl... 75 2e-14
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 172 bits (436), Expect = 9e-44
Identities = 138/484 (28%), Positives = 213/484 (43%), Gaps = 81/484 (16%)
Frame = +1
Query: 2 PFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGS--NVIP 59
P P++G+INP+ L KLL + H++FV TE + P+ + IP
Sbjct: 34 PSPLQGHINPLFQLAKLL--HLRGFHITFVHTEYNHKRLIKSRGPNALDGLPDFVFETIP 207
Query: 60 SELICG-----RDHPAFMEDVMTKMEAPFEELLDLLDH--------PPSIIVYDTLLYWA 106
L G +D + + + K PF++LL L P + +V D L+ +
Sbjct: 208 DGLEDGDGEVSQDLYSLCDSIRNKCHLPFQDLLAKLHRSASAGLVPPVTCLVSDLLMTFT 387
Query: 107 VVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVKYPGF----------EWI--- 153
+ A + +P L P AS F FLH G P+K + +WI
Sbjct: 388 IQAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNGYLDTKVDWIPGM 567
Query: 154 --------------------------------HKAQYLLFSSIYELESQAIDVLKSKLPL 181
+A ++F++ ELE + L + LP
Sbjct: 568 QNFRLKDLPDFIRTTDPNDIMLEFMVEVADRAKRASAIVFNTFNELERDVLSALSTMLP- 744
Query: 182 PIYTIGPTIPKF--------------SLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQ 227
+Y IGP P F +L K D K ++WL+S+ GSV+Y+
Sbjct: 745 SLYPIGP-FPSFLNQAPQNQLASLGSNLWKEDTK--------CLQWLESKEPGSVVYVNF 897
Query: 228 GSFFSVSSAQIDEIAAALCASNVRFLWIAR----SEASRL---KEICGAHHMGLIMEWCD 280
GS +S Q+ E A L FLWI R ++ S + K + + GLI WC
Sbjct: 898 GSITVMSPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPKFVDETSNRGLIASWCP 1077
Query: 281 QLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKE 340
Q +VL+HPS+GGF +HCGWNST ES+ AGVP L P DQP N + + +W +G V
Sbjct: 1078QEKVLNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDIGIEV-- 1251
Query: 341 DVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFI 400
DT VK++++ L+HE M G+ + +R+R+ +L+K GG ++ + + I
Sbjct: 1252----DTNVKREEVENLIHELM--VGDKGKKMRQRTMELKKKAEEDTRPGGCSYMNLDKLI 1413
Query: 401 SDVM 404
+V+
Sbjct: 1414KEVL 1425
>TC12906 similar to PIR|T00584|T00584 indole-3-acetate
beta-glucosyltransferase homolog T27E13.12
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(14%)
Length = 675
Score = 162 bits (410), Expect = 9e-41
Identities = 78/124 (62%), Positives = 96/124 (76%), Gaps = 2/124 (1%)
Frame = +2
Query: 285 LSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPF--NSKMMVEDWKVGCRVKEDV 342
L H ++GGFW+HCGWNSTKE ++AGVPFLT PI+IDQP NSKM+VEDWKVG RVKE V
Sbjct: 2 LCHAAVGGFWTHCGWNSTKEGVLAGVPFLTFPIFIDQPLDSNSKMIVEDWKVGWRVKESV 181
Query: 343 KRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISD 402
+ + VKKD+IV+LV FMDLD + +IRER+KKLQ IC +I GG A +D NAFIS
Sbjct: 182 EVNGFVKKDEIVRLVKRFMDLDSDFAGEIRERAKKLQFICHRAIEIGGFADSDLNAFIST 361
Query: 403 VMHL 406
+M +
Sbjct: 362 IMQV 373
>AV408145
Length = 422
Score = 115 bits (287), Expect = 2e-26
Identities = 62/109 (56%), Positives = 77/109 (69%), Gaps = 5/109 (4%)
Frame = +2
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFI---SSEPK-PDNISFRSGSN 56
+P+P RG++NPMMNL KLL+S N NI ++FVVTEEWL+ I S +PK P N+ F S N
Sbjct: 95 VPYPGRGHVNPMMNLSKLLISKNPNILITFVVTEEWLTLITTSSDDPKPPQNLRFASMPN 274
Query: 57 VIPSELICGRDHPAFMEDVMTKMEAPFEELL-DLLDHPPSIIVYDTLLY 104
VIP E D F+E VMTKMEAPF+ELL LD PPS++VYD L+
Sbjct: 275 VIPPERNRADDFVGFLEAVMTKMEAPFDELLRRQLDPPPSVVVYDAYLF 421
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 110 bits (276), Expect = 3e-25
Identities = 64/169 (37%), Positives = 90/169 (52%), Gaps = 11/169 (6%)
Frame = +2
Query: 248 SNVRFLWIARSEASR-----------LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSH 296
S V FLWI R + L EI G G I WC Q VLSHPSIG F +H
Sbjct: 206 SKVPFLWILRPDVVMGEDSINVPQEFLDEIKGR---GYIASWCFQEEVLSHPSIGAFLTH 376
Query: 297 CGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKL 356
CGWNST E + AG+P + P + +Q NS+ W++G V DVKR D+I L
Sbjct: 377 CGWNSTIEGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHDVKR------DEITTL 538
Query: 357 VHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVMH 405
V+E M GE +++R++ + +K + + GGS++ DF+ I + +H
Sbjct: 539 VNEMM--KGEKGKEMRKKGLEWKKKAIEATGLGGSSYNDFHKLIKESLH 679
>CB829602
Length = 542
Score = 110 bits (276), Expect = 3e-25
Identities = 56/147 (38%), Positives = 88/147 (59%), Gaps = 8/147 (5%)
Frame = +1
Query: 211 IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR-----SEASRL-- 263
++WLDSQ SVLY+ GS +++ Q+ E+A + S +F+W+ R EAS +
Sbjct: 97 LKWLDSQEPSSVLYVNFGSVINMTPQQLVELAWGIANSKKKFIWVIRPDLVEGEASIVLP 276
Query: 264 KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPF 323
+ + G+++ WC Q +VL H ++GGF +HCGWNST ES+ +GVP + P + DQ
Sbjct: 277 EIVAETKDRGIMLSWCPQEQVLKHSALGGFLTHCGWNSTIESISSGVPLICSPFFNDQFV 456
Query: 324 NSKMMVEDWKVGCRV-KEDVKRDTLVK 349
NS+ + +W G + +DVKRD + K
Sbjct: 457 NSRYICSEWDFGMEMNSDDVKRDEVEK 537
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 109 bits (273), Expect = 7e-25
Identities = 105/392 (26%), Positives = 161/392 (40%), Gaps = 76/392 (19%)
Frame = +2
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEW----------------------LSFI 40
FP +G+INP++ L K L + S++ ++ T F
Sbjct: 56 FPAQGHINPLLRLAKCLAAKGSSVTLATTQTAGKSMRTATNITDKAPTPISDGSLKFEFF 235
Query: 41 SSEPKPDNISFRSGSNVIPSEL-ICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVY 99
PD+ S R ++ +L + GR M K A + S I+
Sbjct: 236 DDGLGPDDDSIRGNLSLYLQQLELVGRQQIL----KMIKEHADSNRQI-------SCIIN 382
Query: 100 DTLLYWAVVVANRRNIPAALFWPMPASIFSVF---LHQHIFEQNGHYPVK---------- 146
+ L W V VA + IP+AL W +++F+ + H + + P+
Sbjct: 383 NPFLPWVVDVAEEQGIPSALLWIQSSAVFTAYYSYFHNLVTFPSKEDPLADVQLPSTSIV 562
Query: 147 ---------------YP--------GFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPI 183
YP F+ ++K +L + ELE ID L SK L I
Sbjct: 563 LKHCEIPDFLHPSCTYPFLGTLILEQFKNLNKTFCILVDTYEELEHDFIDYL-SKTFL-I 736
Query: 184 YTIGPTIP----KFSLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQID 239
+GP K + I+ D S+ IEWL+S+P SV+Y++ GS + Q+D
Sbjct: 737 RPVGPLFKSSQAKSAAIRGDFMKSDD----CIEWLNSKPQSSVVYVSFGSIVYLPQEQVD 904
Query: 240 EIAAALCASNVRFLWIARSEASRLKEICGAH-------------HMGLIMEWCDQLRVLS 286
EIA L S V FLW+ + E+ G G +++W Q VL+
Sbjct: 905 EIAFGLKKSQVSFLWVMKPPP----EVTGLQPHVLPYGFLEETGERGRVVQWSPQEEVLA 1072
Query: 287 HPSIGGFWSHCGWNSTKESLVAGVPFLTLPIY 318
HPS+ F +HCGWNS+ E+L +GVP LT P +
Sbjct: 1073HPSVSCFLTHCGWNSSMEALTSGVPVLTFPAW 1168
>CN825072
Length = 712
Score = 106 bits (264), Expect = 8e-24
Identities = 69/203 (33%), Positives = 103/203 (49%), Gaps = 25/203 (12%)
Frame = -3
Query: 228 GSFFSVSSAQIDEIAAALCASNVRFLWIARSE-----------------ASRLKE--ICG 268
GS S AQI EIA A+ S VRF+W R AS L E +
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDR 531
Query: 269 AHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMM 328
+G ++ W Q RVL+HP+ GGF SHCGWNST ES+ GVP T P+Y +Q N+ ++
Sbjct: 530 TTGIGRVIGWAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFVL 351
Query: 329 VEDWKVGCRVKEDVKRDT------LVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKIC 382
V + K+ + D + + L+ DKI + +D DGE+ + ++E S+K +K
Sbjct: 350 VRELKIAVEISLDYRVEINGGPNYLLTADKIEGGIRSVLDKDGEVRKRVKEMSEKSRK-- 177
Query: 383 LNSIANGGSAHTDFNAFISDVMH 405
++ GG +++ + I MH
Sbjct: 176 --TLLEGGCSYSYLDRLIDYFMH 114
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 105 bits (263), Expect = 1e-23
Identities = 66/221 (29%), Positives = 114/221 (50%), Gaps = 18/221 (8%)
Frame = +1
Query: 152 WIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSYYI 211
W++ +L F+++ + +S + + KL P + IGP + K +
Sbjct: 211 WVNSDGFL-FNTVADFDSMGLRYVARKLNRPAWAIGPVLLSTGSGSRG-KGGGISPELCR 384
Query: 212 EWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR-------SEASRLK 264
+WLD++P SVL+++ GS ++S++Q+ ++A AL S F+W+ R + R +
Sbjct: 385 KWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEFRAE 564
Query: 265 EICGAHHMG-------LIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPI 317
E +G ++ +W Q+ +LSH ++ F SHCGWNS ESL GVP L P+
Sbjct: 565 EWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILGWPM 744
Query: 318 YIDQPFNSKMMVEDWKVGCRVKE----DVKRDTLVKKDKIV 354
+Q FN KM+ E+ V V +V+ + LV+K ++V
Sbjct: 745 AAEQFFNCKMLEEELGVCVEVARGKRCEVRHEDLVEKIELV 867
>TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (26%)
Length = 565
Score = 103 bits (256), Expect = 7e-23
Identities = 51/132 (38%), Positives = 80/132 (59%)
Frame = +1
Query: 273 GLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDW 332
GLI WC Q VL+HPSIGGF +HCGWNST ES+ AGVP L P + DQ N + + +W
Sbjct: 52 GLIASWCSQEHVLNHPSIGGFLTHCGWNSTIESISAGVPMLCWPFFADQLTNCRSICNEW 231
Query: 333 KVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
+G ++ DT K++++ KL++E M GE + +R+++ +L+K GG +
Sbjct: 232 DIGMQI------DTNGKREEVEKLINELM--VGEKGKKMRQKTMELKKRAEEDTRPGGCS 387
Query: 393 HTDFNAFISDVM 404
+ + + I +V+
Sbjct: 388 YMNLDQVIKEVL 423
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 102 bits (253), Expect = 2e-22
Identities = 63/188 (33%), Positives = 94/188 (49%), Gaps = 20/188 (10%)
Frame = +3
Query: 211 IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR--SEASRLKEICG 268
+ WLD QP GSV+Y++ GS ++ Q+ EIA L S RF+W+ R +EA G
Sbjct: 63 LRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQHRFVWVVRPPNEADASATFFG 242
Query: 269 -----------------AHHMGLIME-WCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGV 310
G+++ W Q +L HP+ GGF + CGWNS ES++ GV
Sbjct: 243 FGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEILGHPATGGFVTPCGWNSVLESVLNGV 422
Query: 311 PFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRD 370
P + P+Y +Q N+ M+ E+ V RVK V +V +D++V +V M + +
Sbjct: 423 PMVAWPLYSEQKMNAYMLSEELGVAVRVK--VAEGGVVCRDQVVDMVRRVMVSEEGMGMK 596
Query: 371 IRERSKKL 378
R R KL
Sbjct: 597 DRVRELKL 620
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 99.0 bits (245), Expect = 1e-21
Identities = 97/370 (26%), Positives = 152/370 (40%), Gaps = 74/370 (20%)
Frame = +1
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNIS------FRSG 54
+P+P +G+INPM+ L KLL HV+FV TE + PD+++ F +
Sbjct: 103 IPYPAQGHINPMLKLAKLL-HFKGGFHVTFVNTEYNHKRLLKSRGPDSLNGLPSFRFETI 279
Query: 55 SNVIP-SELICGRDHPAFMEDVMTKMEAPFEELLDLL-----DHPP-SIIVYDTLLYWAV 107
+ +P +++ +D P+ F++LL L D PP + IV D + + +
Sbjct: 280 PDGLPETDVDVTQDIPSLCISTRKTCLPHFKKLLSKLNDVSSDVPPVTCIVSDGCMSFTL 459
Query: 108 VVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVK----------------YPGFE 151
A NIP LFW A F ++ + G P+K PG +
Sbjct: 460 DAAIELNIPEVLFWTTSACGFMGYVQYRELIEKGIIPLKDSSDITNGYLETTIEWLPGMK 639
Query: 152 WIH-----------------------------KAQYLLFSSIYELESQAIDVLKSKLPLP 182
I KA ++ ++ LE ++ S LP P
Sbjct: 640 NIRLKDLPSFLRTTDPNDKMLDFLTGECQRALKASAIILNTFDALEHDVLEAFSSILP-P 816
Query: 183 IYTIGPTIPKFSLIKNDPKPSNTNH---------SYYIEWLDSQPIGSVLYIAQGSFFSV 233
+Y+IGP L+ D N N S ++WLD++ SV+Y+ GS +
Sbjct: 817 VYSIGP----LHLLIKDVTDKNLNSLGSNLWKEDSECLKWLDTKEPNSVVYVNFGSITVM 984
Query: 234 SSAQIDEIAAALCASNVRFLWIARSEASRLKE-------ICGAHHMGLIMEWCDQLRVLS 286
+S Q+ E A L SN FLW+ R + K + + G + W Q VL+
Sbjct: 985 TSEQMVEFAWGLANSNKTFLWVIRPDLVAGKHAVLPEEFVAATNDRGRLSSWTPQEDVLT 1164
Query: 287 HPSIGGFWSH 296
HP+IGGF +H
Sbjct: 1165HPAIGGFLTH 1194
>BI421006
Length = 519
Score = 89.0 bits (219), Expect(2) = 2e-21
Identities = 51/134 (38%), Positives = 73/134 (54%), Gaps = 15/134 (11%)
Frame = +2
Query: 230 FFSVSSAQIDEIAAALCASNVRFLWIARS-------EASRLKEIC-------GAHHMGLI 275
F + QI++IA L S +F+W+ R + ++KE MGL+
Sbjct: 110 FSTFIEEQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGFEKRVEGMGLV 289
Query: 276 M-EWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKV 334
+ +W QL +LSHPS GGF SHCGWNS ES+ GVP P++ DQP N+ ++ E KV
Sbjct: 290 VRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEVLKV 469
Query: 335 GCRVKEDVKRDTLV 348
V++ +RD LV
Sbjct: 470 ALVVQDXAQRDELV 511
Score = 29.6 bits (65), Expect(2) = 2e-21
Identities = 13/30 (43%), Positives = 19/30 (63%)
Frame = +3
Query: 201 KPSNTNHSYYIEWLDSQPIGSVLYIAQGSF 230
K S+ ++ +EWLD Q SVLY++ G F
Sbjct: 24 KNSSMGRNFVMEWLDRQEQKSVLYVSFGFF 113
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 96.7 bits (239), Expect = 6e-21
Identities = 47/133 (35%), Positives = 77/133 (57%)
Frame = +2
Query: 273 GLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDW 332
GLI+ WC QL VL H ++G F +HCGWNST ES+ GVP + +P++ D N K++V+ W
Sbjct: 26 GLIVTWCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPVTNGKLIVDVW 205
Query: 333 KVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
K G R D K +V+++ I + E ++ E +I+ + K + +S+A GG +
Sbjct: 206 KTGVRAVADEKE--IVRRETIKNCIKEI--IETEKINEIKSNAIKWMNLAKDSLAEGGRS 373
Query: 393 HTDFNAFISDVMH 405
+ F++ + H
Sbjct: 374 DKNIAEFVAALDH 412
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 91.7 bits (226), Expect = 2e-19
Identities = 45/134 (33%), Positives = 76/134 (56%)
Frame = +1
Query: 273 GLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDW 332
G ++W Q +VL HP+IG FW+HCGWNST ES+ GVP + P + DQ N+K + + W
Sbjct: 7 GTSVKWAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSDVW 186
Query: 333 KVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
KVG +++ R + K ++ L E ++ G + + K+ +CL + GGS+
Sbjct: 187 KVGVQLQNKPGRGEIEKTISLLMLGDEAYEIRGNIL-----KLKEKANVCL---SEGGSS 342
Query: 393 HTDFNAFISDVMHL 406
+ + +S+++ L
Sbjct: 343 YCFLDRLVSEILSL 384
>TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (19%)
Length = 571
Score = 90.9 bits (224), Expect = 3e-19
Identities = 46/133 (34%), Positives = 76/133 (56%)
Frame = +1
Query: 273 GLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDW 332
G +++W Q ++L+HP+I F SHCGWNST E + A VPFL P DQ N + + W
Sbjct: 16 GKVVKWGPQKKILNHPAIACFISHCGWNSTIEGVHASVPFLCWPFSTDQFLNKSFICDVW 195
Query: 333 KVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
K+G + +D + ++ K +I V + + + D++ RS KL++I +N++A GG +
Sbjct: 196 KIGLGLDKD--ENGIIPKGEIRTKVEQVI-----VNEDLKARSLKLKEITINNLAEGGQS 354
Query: 393 HTDFNAFISDVMH 405
+ FI+ H
Sbjct: 355 SNNLKKFINWTKH 393
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 90.5 bits (223), Expect = 5e-19
Identities = 48/140 (34%), Positives = 77/140 (54%), Gaps = 7/140 (5%)
Frame = +1
Query: 274 LIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWK 333
+I W Q+ +L H ++G F +HCGWNST E++ AGVP +T P+ +Q +N K++ E
Sbjct: 16 IIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLITEVRG 195
Query: 334 VGCRVKEDV-------KRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSI 386
+G V + KR+ LV ++ I K V MD G+ IR R+++ + ++
Sbjct: 196 IGVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMD-GGDEGEQIRRRAQEYGEKARQAV 372
Query: 387 ANGGSAHTDFNAFISDVMHL 406
GGS+H + A I D+ L
Sbjct: 373 EEGGSSHKNLTALIDDLKRL 432
>BP039584
Length = 545
Score = 90.1 bits (222), Expect = 6e-19
Identities = 46/135 (34%), Positives = 76/135 (56%), Gaps = 6/135 (4%)
Frame = -3
Query: 278 WCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCR 337
W QL +L HP+IGG +HCGWN+T ES++AG+P T+P++ +Q +N K++V+ VG
Sbjct: 537 WAPQLLILEHPAIGGVVTHCGWNTTLESVIAGLPMATMPLFAEQFYNEKLLVDVLGVGVS 358
Query: 338 V------KEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGS 391
+ +V D +VK++ IVK + M GE ++R R ++L +I GGS
Sbjct: 357 IGMKKWRNWNVVGDEIVKRENIVKAISLLMG-GGEEALEMRRRVRELSDAAKKTIQPGGS 181
Query: 392 AHTDFNAFISDVMHL 406
++ ++ L
Sbjct: 180 SYNKVKGLFDELKAL 136
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 89.4 bits (220), Expect = 1e-18
Identities = 50/135 (37%), Positives = 74/135 (54%), Gaps = 5/135 (3%)
Frame = +1
Query: 274 LIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWK 333
+I W Q+ +L H +IG F +HCGWNST E + AGVP +T PI +Q +N K++ E K
Sbjct: 1 IIRGWAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLK 180
Query: 334 VGCRVKED-----VKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIAN 388
+G V V VK D + K V M +GE ++R R K L + ++
Sbjct: 181 IGVPVGAKKWGRMVLEGDGVKSDAVEKGVKRIM--EGEEAEEMRNRVKVLSQQAQWAVEE 354
Query: 389 GGSAHTDFNAFISDV 403
GGS+++D NA I ++
Sbjct: 355 GGSSYSDLNALIEEL 399
>TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (26%)
Length = 643
Score = 80.9 bits (198), Expect = 4e-16
Identities = 45/112 (40%), Positives = 66/112 (58%)
Frame = +3
Query: 281 QLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKE 340
Q +VLSH SIGGF SHCGWNS ES+ GVP + P+Y +Q N+ ++ ED KV R K
Sbjct: 9 QAKVLSHASIGGFLSHCGWNSVLESVANGVPLVAWPLYAEQRMNAVLVTEDVKVALRPK- 185
Query: 341 DVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSA 392
V + LV+ +I ++V M GE + +R + K L++ ++ GS+
Sbjct: 186 -VGENGLVELVEIARVVRTLM--QGEEGKKLRCKMKDLKEAAATTLKENGSS 332
>TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (19%)
Length = 542
Score = 75.5 bits (184), Expect = 2e-14
Identities = 44/137 (32%), Positives = 71/137 (51%), Gaps = 8/137 (5%)
Frame = +2
Query: 278 WCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCR 337
W Q +L+H + GGF +HCGWN+ E++ AGVP +T+P + DQ +N K++ E G
Sbjct: 8 WAPQPLILNHLATGGFLTHCGWNAVVEAISAGVPMITMPGFSDQYYNEKLITEVHGFGVE 187
Query: 338 V--------KEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANG 389
V D K+ +V ++I K V M GE +IR ++K++Q ++ G
Sbjct: 188 VGAAEWSISPYDGKK-KVVSGERIEKAVKSLM---GEEGAEIRSKAKEVQDKAWKAVQQG 355
Query: 390 GSAHTDFNAFISDVMHL 406
GS++ I + L
Sbjct: 356 GSSYNSLTVLIDHLRTL 406
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,530,087
Number of Sequences: 28460
Number of extensions: 135734
Number of successful extensions: 745
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 723
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 729
length of query: 406
length of database: 4,897,600
effective HSP length: 92
effective length of query: 314
effective length of database: 2,279,280
effective search space: 715693920
effective search space used: 715693920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC141923.7


