

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.4 + phase: 0 /partial
(1196 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
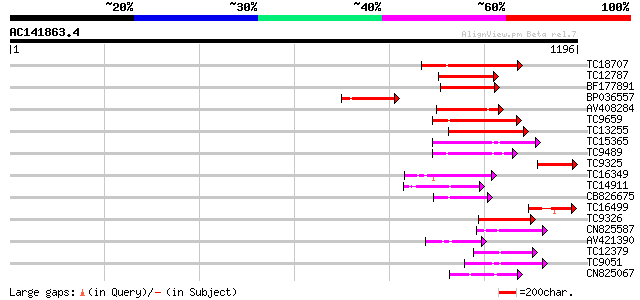
Score E
Sequences producing significant alignments: (bits) Value
TC18707 homologue to PIR|T02901|T02901 MSP1 protein homolog T13J... 300 9e-82
TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (T... 241 4e-64
BF177891 180 1e-45
BP036557 179 2e-45
AV408284 173 1e-43
TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATP... 169 2e-42
TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%) 160 8e-40
TC15365 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {... 147 1e-35
TC9489 homologue to UP|Q9SEI5 (Q9SEI5) 26S proteasome AAA-ATPase... 121 6e-28
TC9325 114 9e-26
TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulat... 106 2e-23
TC14911 homologue to UP|Q93X55 (Q93X55) Peroxin 6 (Fragment), pa... 102 5e-22
CB826675 94 1e-19
TC16499 similar to UP|Q7X9J7 (Q7X9J7) P60 katanin (Fragment), pa... 92 4e-19
TC9326 homologue to UP|Q93WX4 (Q93WX4) Suppressor of K+ transpor... 89 3e-18
CN825587 88 7e-18
AV421390 82 5e-16
TC12379 homologue to UP|O04327 (O04327) Cell division protein Ft... 79 4e-15
TC9051 UP|PRS7_ARATH (Q9SSB5) 26S protease regulatory subunit 7 ... 78 7e-15
CN825067 75 5e-14
>TC18707 homologue to PIR|T02901|T02901 MSP1 protein homolog T13J8.110 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(29%)
Length = 771
Score = 300 bits (768), Expect = 9e-82
Identities = 147/214 (68%), Positives = 181/214 (83%)
Frame = +3
Query: 868 KDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLT 927
K V +NEFEKR+ +VIP N+IGVTF DIGA++ +K++L+ELVMLPL+RP+LF KG L
Sbjct: 120 KAEVPDNEFEKRIRPEVIPANEIGVTFGDIGAMDEIKESLQELVMLPLRRPDLF-KGGLL 296
Query: 928 KPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 987
KPC+GILLFGPPGTGKTMLA A+A +AGA+FIN+SMS+ITSKWFGE EK V+A+FSLA+K
Sbjct: 297 KPCRGILLFGPPGTGKTMLAXAIANEAGASFINVSMSTITSKWFGEDEKNVRALFSLAAK 476
Query: 988 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYD 1047
+AP++IFVDEVDSMLG+R GEHEAMRK+KNEFM +WDGL T E+++VLAATNRP+D
Sbjct: 477 VAPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTAPGEQILVLAATNRPFD 656
Query: 1048 LDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKE 1081
LDEA+IRR RR+MV LP NR ILK +LA E
Sbjct: 657 LDEAIIRRFERRIMVGLPSVENREMILKTLLA*E 758
>TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (TAT-binding
homolog 4), partial (20%)
Length = 389
Score = 241 bits (616), Expect = 4e-64
Identities = 118/128 (92%), Positives = 124/128 (96%)
Frame = +3
Query: 904 KDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISM 963
+DTLKELVMLPL+RPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT+AGANFINISM
Sbjct: 3 EDTLKELVMLPLKRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISM 182
Query: 964 SSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMV 1023
SSITSKWFGEGEKYVKAVFSLASKI+PSVIFVDEVD ML RRENPGEHEAMRKM NEFMV
Sbjct: 183 SSITSKWFGEGEKYVKAVFSLASKISPSVIFVDEVDXMLXRRENPGEHEAMRKMXNEFMV 362
Query: 1024 NWDGLRTK 1031
+WDGL +K
Sbjct: 363 HWDGLPSK 386
>BF177891
Length = 390
Score = 180 bits (456), Expect = 1e-45
Identities = 84/124 (67%), Positives = 105/124 (83%)
Frame = +1
Query: 909 ELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITS 968
ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AKA+AT+AGANFI++S S++TS
Sbjct: 16 ELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTS 195
Query: 969 KWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGL 1028
KWFG+ EK KA+FS ASK+AP +IFVDEVDS+LG R EHEA R+M+NEFM WDGL
Sbjct: 196 KWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGL 375
Query: 1029 RTKD 1032
R+K+
Sbjct: 376 RSKE 387
>BP036557
Length = 369
Score = 179 bits (455), Expect = 2e-45
Identities = 87/123 (70%), Positives = 100/123 (80%)
Frame = +2
Query: 700 KSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQD 759
KSH GG + + L FP F ++HDRGK+VPKPNKTLTKLFPNKVTIHMPQD
Sbjct: 2 KSHPGGCFLRSW*QSDCPS*-LGFPR*FRQMHDRGKDVPKPNKTLTKLFPNKVTIHMPQD 178
Query: 760 EALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKIL 819
E LLASWK QLDRDVETLKIKGNLHHLRTVLSR GME +GL++LC+KD T+TNEN+EK++
Sbjct: 179 ETLLASWKDQLDRDVETLKIKGNLHHLRTVLSRCGMECEGLDTLCIKDQTVTNENAEKVV 358
Query: 820 GWA 822
GWA
Sbjct: 359 GWA 367
>AV408284
Length = 425
Score = 173 bits (439), Expect = 1e-43
Identities = 84/142 (59%), Positives = 110/142 (77%)
Frame = +1
Query: 900 LENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFI 959
LE++K+ L ELV+LPL+RP+LF G+L P KG+LL+GPPGTGKTMLAKA+A ++GA FI
Sbjct: 1 LESIKEALFELVILPLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFI 180
Query: 960 NISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKN 1019
N+ +S++ SKWFG+ +K V AVFSLA K+ P++IF+DEVDS LG+R +HEA+ MK
Sbjct: 181 NVRISNLMSKWFGDAQKLVAAVFSLAHKLQPAIIFIDEVDSFLGQRRTT-DHEALLNMKT 357
Query: 1020 EFMVNWDGLRTKDTERVIVLAA 1041
EFM WDG T RV+VLAA
Sbjct: 358 EFMALWDGFTTDQNARVMVLAA 423
>TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase, partial
(74%)
Length = 1060
Score = 169 bits (428), Expect = 2e-42
Identities = 85/187 (45%), Positives = 126/187 (66%)
Frame = +1
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
+ ++D+ LE+ K +L+E V+LP++ P+ F + +P + LL+GPPGTGK+ LAKAVA
Sbjct: 454 IKWNDVAGLESAKQSLQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 627
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
T+A + F ++S S + SKW GE EK V +F +A + APS+IFVDE+DS+ G+R E
Sbjct: 628 TEADSTFFSVSSSDLGSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQRGEGNES 807
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRA 1071
EA R++K E +V G+ D ++V+VLAATN PY LD+A+ RR +R+ + LPD R
Sbjct: 808 EASRRIKTELLVQMQGVGNND-QKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDVKARQ 984
Query: 1072 KILKVIL 1078
+ KV L
Sbjct: 985 HMFKVHL 1005
>TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%)
Length = 532
Score = 160 bits (406), Expect = 8e-40
Identities = 84/170 (49%), Positives = 113/170 (66%), Gaps = 1/170 (0%)
Frame = +1
Query: 926 LTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLA 985
L P KGILLFGPPGTGKTMLAKAVAT+ F NIS SS+ SKW G+ EK VK +F LA
Sbjct: 22 LLSPWKGILLFGPPGTGKTMLAKAVATECKTTFFNISASSVVSKWRGDSEKLVKVLFQLA 201
Query: 986 SKIAPSVIFVDEVDSMLGRR-ENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNR 1044
APS IF+DE+D+++ +R E EHEA R++K E ++ DGL T+ E V VLAATN
Sbjct: 202 RHHAPSTIFLDEIDAIISQRGEARSEHEASRRLKTELLIQMDGL-TRTDELVFVLAATNL 378
Query: 1045 PYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIAN 1094
P++LD A++RRL +R++V LP+ R + + +L + + + N
Sbjct: 379 PWELDAAMLRRLEKRILVPLPEPEARVAMFEELLPPQPDEESIPYDLLVN 528
>TC15365 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {Arabidopsis
thaliana;}, partial (87%)
Length = 1385
Score = 147 bits (371), Expect = 1e-35
Identities = 89/232 (38%), Positives = 137/232 (58%), Gaps = 6/232 (2%)
Frame = +3
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT 952
T+D IG L+ +KE++ LP++ PELF + +P KG+LL+GPPGTGKT+LA+AVA
Sbjct: 321 TYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQP-KGVLLYGPPGTGKTLLARAVAH 497
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR----ENP 1008
FI +S S + K+ GEG + V+ +F +A + APS+IF+DE+DS+ R
Sbjct: 498 HTDCTFIRVSGSELVQKYIGEGSRMVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGX 677
Query: 1009 GEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPD 1066
G+ E R M E + DG + ++ VL ATNR LD+A++R R+ R++ P+
Sbjct: 678 GDSEVQRTML-ELLNQLDGFEA--SNKIKVLMATNRIDILDQALLRPGRIDRKIEFPNPN 848
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE 1118
+R ILK+ + +L +DL IA +G SG++LK +C A ++E
Sbjct: 849 EESRQDILKIHSRRMNLMRGIDLKKIAEKMNGASGAELKAVCTEAGMFALRE 1004
>TC9489 homologue to UP|Q9SEI5 (Q9SEI5) 26S proteasome AAA-ATPase subunit
RPT2a, partial (69%)
Length = 916
Score = 121 bits (304), Expect = 6e-28
Identities = 72/183 (39%), Positives = 110/183 (59%), Gaps = 5/183 (2%)
Frame = +1
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVAT 952
++ DIG L+ +KE V LPL PEL+ + KP KG++L+G PGTGKT+LAKAVA
Sbjct: 376 SYADIGGLDAQIQEIKEAVELPLTHPELY-EDIGIKPPKGVILYGEPGTGKTLLAKAVAN 552
Query: 953 DAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR---ENPG 1009
A F+ + S + K+ G+G K V+ +F +A ++PS++F+DE+D++ +R + G
Sbjct: 553 STSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGG 732
Query: 1010 EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
E E R M E + DG ++ +VI+ ATNR LD A++R R+ R++ LPD
Sbjct: 733 EREIQRTML-ELLNQLDGFDSRGDVKVIL--ATNRIESLDPALLRPGRIDRKIEFPLPDT 903
Query: 1068 PNR 1070
R
Sbjct: 904 KTR 912
>TC9325
Length = 672
Score = 114 bits (285), Expect = 9e-26
Identities = 55/84 (65%), Positives = 67/84 (79%)
Frame = +2
Query: 1113 HRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNM 1172
H PI+E+LEKEKKE + A+AE P P L S DIR+L MEDF++AH+QV ASVSS+S NM
Sbjct: 2 HCPIREVLEKEKKERSLALAENEPLPGLCSSADIRALKMEDFRYAHEQVGASVSSDSTNM 181
Query: 1173 TELVQWNELYGEGGSRVKKALSYF 1196
EL QWN+LYGEGGSR ++LSYF
Sbjct: 182 NELQQWNDLYGEGGSRKVRSLSYF 253
>TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulatory subunit
6B homolog, partial (65%)
Length = 901
Score = 106 bits (264), Expect = 2e-23
Identities = 74/209 (35%), Positives = 107/209 (50%), Gaps = 14/209 (6%)
Frame = +3
Query: 833 ADADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIG- 891
AD + +V S+ Y + I I E LK S + + L DV+PP
Sbjct: 291 ADQNNGIVGSTTGSNYYVRILSTINREL--LKPSASVALHRHS---NALVDVLPPEADSS 455
Query: 892 -----------VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPG 940
VT++DIG + K ++E V LPL EL+ + + P +G+LL+GPPG
Sbjct: 456 ISLLSQSEKPDVTYNDIGGCDIQKQEIREAVELPLTHHELYKQIGIDPP-RGVLLYGPPG 632
Query: 941 TGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDS 1000
TGKTMLAKAVA A FI + S K+ GEG + V+ VF LA + AP++IF+DEVD+
Sbjct: 633 TGKTMLAKAVANHTTAAFIRVVGSEFVQKYLGEGPRMVRDVFRLAKENAPAIIFIDEVDA 812
Query: 1001 MLGRR--ENPGEHEAMRKMKNEFMVNWDG 1027
+ R G ++++ E + DG
Sbjct: 813 IATARFDAQTGADREVQRILMELLNQMDG 899
>TC14911 homologue to UP|Q93X55 (Q93X55) Peroxin 6 (Fragment), partial (32%)
Length = 1418
Score = 102 bits (253), Expect = 5e-22
Identities = 65/177 (36%), Positives = 97/177 (54%), Gaps = 5/177 (2%)
Frame = +2
Query: 830 NPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKK----SLKDVVTENEFEKRLLGDVI 885
N E D D + S S+ GI I++ +KS +DV+ E K+ +
Sbjct: 32 NAEVDKDGSEDVDS-SLTSGI-----IEDNNKSKVSPRIPGKEDVMNALERSKKRNASAL 193
Query: 886 -PPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKT 944
P V ++D+G LE+VK ++ + V LPL +LF G + G+LL+GPPGTGKT
Sbjct: 194 GTPKVPNVKWEDVGGLEDVKKSILDTVQLPLLHKDLFASGLRKR--SGVLLYGPPGTGKT 367
Query: 945 MLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSM 1001
+LAKAVAT+ NF+++ + + + GE EK V+ +F A P VIF DE+DS+
Sbjct: 368 LLAKAVATECSLNFLSVKGPELINMYIGESEKNVRDIFQKARSARPCVIFFDELDSL 538
>CB826675
Length = 551
Score = 94.4 bits (233), Expect = 1e-19
Identities = 48/127 (37%), Positives = 77/127 (59%), Gaps = 3/127 (2%)
Frame = +1
Query: 894 FDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATD 953
++D+G LE L E ++LP+ E F K + +P KG+LL+GPPGTGKT++A+A A
Sbjct: 133 YNDVGGLEKQIQELVEAIVLPMTHKERFQKLGV-RPPKGVLLYGPPGTGKTLMARACAAQ 309
Query: 954 AGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRR---ENPGE 1010
A F+ ++ + + G+G K V+ F LA + +P +IF+DE+D++ +R E G+
Sbjct: 310 TNATFLKLAGPQLVQMFIGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKRFDSEVSGD 489
Query: 1011 HEAMRKM 1017
E R M
Sbjct: 490 REVQRTM 510
>TC16499 similar to UP|Q7X9J7 (Q7X9J7) P60 katanin (Fragment), partial (61%)
Length = 646
Score = 92.4 bits (228), Expect = 4e-19
Identities = 47/107 (43%), Positives = 69/107 (63%), Gaps = 5/107 (4%)
Frame = +1
Query: 1094 NMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDI-----RS 1148
N+TDGYSGSDLKNLC+TAA+RP++E+LE+EK G ++I R
Sbjct: 1 NLTDGYSGSDLKNLCITAAYRPVQELLEEEK----------------AGDNNITNVALRP 132
Query: 1149 LNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
LN++DF + QV SV+ ++V++ EL +WNE+YGEGG+R + +
Sbjct: 133 LNLDDFVQSKSQVGPSVAYDAVSVNELRKWNEMYGEGGTRTQSPFGF 273
>TC9326 homologue to UP|Q93WX4 (Q93WX4) Suppressor of K+ transport growth
defect-like protein (Fragment), partial (73%)
Length = 894
Score = 89.4 bits (220), Expect = 3e-18
Identities = 50/121 (41%), Positives = 76/121 (62%), Gaps = 1/121 (0%)
Frame = +2
Query: 990 PSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLD 1049
P FVDE+DS+ G+R E EA R++K E +V G+ D ++V+VLAATN PY LD
Sbjct: 8 PFYHFVDEIDSLCGQRGEGNESEASRRIKTELLVQMQGVGNND-QKVLVLAATNTPYALD 184
Query: 1050 EAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS-SDVDLGAIANMTDGYSGSDLKNLC 1108
+A+ RR +R+ + LPD R + KV L + ++ D +A+ T+G+SGSD+ ++C
Sbjct: 185 QAIRRRFDKRIYIPLPDLKARQHMFKVHLGDTPHNLNESDFEYLASRTEGFSGSDI-SVC 361
Query: 1109 V 1109
V
Sbjct: 362 V 364
>CN825587
Length = 663
Score = 88.2 bits (217), Expect = 7e-18
Identities = 58/159 (36%), Positives = 87/159 (54%), Gaps = 9/159 (5%)
Frame = +2
Query: 985 ASKIAPSVIFVDEVDSMLGRRENPG---EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAA 1041
A AP ++F+DE+D++ GR+ G ++ + N+ + DG VIVLAA
Sbjct: 2 AKSKAPCIVFIDEIDAV-GRQRGAGLGGGNDEREQTINQLLTEMDGF--SGNSGVIVLAA 172
Query: 1042 TNRPYDLDEAVIR--RLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGY 1099
TNRP LD A++R R R++ V+ PD R KIL+V + L+ DVD IA T G+
Sbjct: 173 TNRPDVLDSALLRPGRFDRQVTVDRPDVAGRVKILQVHSRGKALAKDVDFDKIARRTPGF 352
Query: 1100 SGSDLKNL----CVTAAHRPIKEILEKEKKELAAAVAEG 1134
+G+DL+NL + AA R +KEI + E + + G
Sbjct: 353 TGADLQNLMNEAAILAARRDLKEISKDEISDALERIIAG 469
>AV421390
Length = 409
Score = 82.0 bits (201), Expect = 5e-16
Identities = 51/131 (38%), Positives = 78/131 (58%), Gaps = 2/131 (1%)
Frame = +2
Query: 878 KRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCK--GQLTKPCKGILL 935
K L +V+P ++ TF D+ ++ K L+E+V L+ P F + G+L K GILL
Sbjct: 23 KELNKEVMPEKNVK-TFKDVKGCDDAKQELEEVVEY-LRNPAKFTRLGGKLPK---GILL 187
Query: 936 FGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFV 995
G PGTGKT+LAKA+A +AG F + S + G G + V+++F A K AP +IF+
Sbjct: 188 TGAPGTGKTLLAKAIAGEAGVPFFYRAGSEFEEMFVGVGARRVRSLFQAAKKKAPCIIFI 367
Query: 996 DEVDSMLGRRE 1006
DE+D++ R+
Sbjct: 368 DEIDAVGSTRK 400
>TC12379 homologue to UP|O04327 (O04327) Cell division protein FtsH isolog,
partial (15%)
Length = 505
Score = 79.0 bits (193), Expect = 4e-15
Identities = 51/139 (36%), Positives = 81/139 (57%), Gaps = 5/139 (3%)
Frame = +1
Query: 979 KAVFSLASKIAPSVIFVDEVDSMLGRR---ENPGEHEAMRKMKNEFMVNWDGLRTKDTER 1035
+A++ A + APSV+F+DE+D++ R + G E + N+ +V DG +
Sbjct: 61 RALYQEAKENAPSVVFIDELDAVGRERGLIKGSGGQERDATL-NQLLVCLDGFEGRG--E 231
Query: 1036 VIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIA 1093
VI +A+TNRP LD A++R R R++ + P R +ILKV K+ ++ DVD A+A
Sbjct: 232 VITIASTNRPDILDPALVRPGRFDRKIFIPKPGLIGRIEILKVHARKKPIAGDVDYMAVA 411
Query: 1094 NMTDGYSGSDLKNLCVTAA 1112
+MTDG G++L N+ AA
Sbjct: 412 SMTDGMVGAELANIVEVAA 468
>TC9051 UP|PRS7_ARATH (Q9SSB5) 26S protease regulatory subunit 7 (26S
proteasome subunit 7) (26S proteasome AAA-ATPase subunit
RPT1a) (Regulatory particle triple-A ATPase subunit 1a),
partial (46%)
Length = 867
Score = 78.2 bits (191), Expect = 7e-15
Identities = 54/185 (29%), Positives = 93/185 (50%), Gaps = 9/185 (4%)
Frame = +2
Query: 959 INISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP---GEHEAMR 1015
I + S + K+ GEG + V+ +F +A ++F DEVD++ G R + G++E R
Sbjct: 2 IRVIGSELVQKYVGEGARMVRELFQMARSKKACIVFFDEVDAIGGARFDDGVGGDNEVQR 181
Query: 1016 KMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPNRAKI 1073
M E + DG + + VL ATNRP LD A++R RL R++ LPD +R +I
Sbjct: 182 TML-EIVNQLDGFDARG--NIKVLMATNRPDTLDPALLRPGRLDRKVEFGLPDLESRTQI 352
Query: 1074 LKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPI----KEILEKEKKELAA 1129
K+ + D+ ++ + +G+D++++C A I K + EK+ +
Sbjct: 353 FKIHTRTMNCERDIRFELLSRLCPNSTGADIRSVCTEAGMYAIRARRKTVTEKDFLDAVN 532
Query: 1130 AVAEG 1134
V +G
Sbjct: 533 KVIKG 547
>CN825067
Length = 687
Score = 75.5 bits (184), Expect = 5e-14
Identities = 53/154 (34%), Positives = 77/154 (49%), Gaps = 2/154 (1%)
Frame = +1
Query: 929 PCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKI 988
P + +L +GPPGTGKTM A+ +A +G ++ ++ + + + +F + K
Sbjct: 187 PFRNMLFYGPPGTGKTMAARELARKSGLDYALMTGGDVAPLG-SQAVTKIHELFDWSKKS 363
Query: 989 APSVI-FVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLA-ATNRPY 1046
++ F+DE D+ L R EA R N + RT D + IVLA ATNRP
Sbjct: 364 KRGLLLFIDEADAFLCERNKTYMSEAQRSALNALL-----FRTGDQSKDIVLALATNRPG 528
Query: 1047 DLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAK 1080
DLD AV R+ L LP R K+LK+ L K
Sbjct: 529 DLDSAVADRIDEVLEFPLPGEGERYKLLKLYLDK 630
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,303,997
Number of Sequences: 28460
Number of extensions: 265635
Number of successful extensions: 1502
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 1381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1464
length of query: 1196
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1095
effective length of database: 2,023,140
effective search space: 2215338300
effective search space used: 2215338300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC141863.4


