

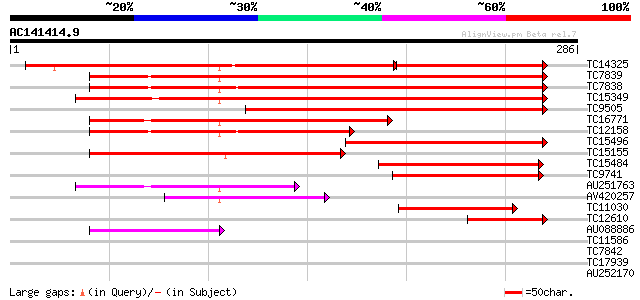
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.9 + phase: 0
(286 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14325 homologue to UP|Q9FXL2 (Q9FXL2) Vf14-3-3c protein, parti... 219 6e-81
TC7839 homologue to UP|Q93XW1 (Q93XW1) 14-3-3 protein, complete 259 5e-70
TC7838 homologue to UP|Q93XW1 (Q93XW1) 14-3-3 protein, complete 256 3e-69
TC15349 homologue to UP|143C_SOYBN (Q96452) 14-3-3-like protein ... 225 6e-60
TC9505 similar to PIR|F86391|F86391 T1K7.15 protein - Arabidopsi... 172 8e-44
TC16771 homologue to UP|O49152 (O49152) 14-3-3 protein homolog, ... 142 5e-35
TC12158 homologue to UP|143A_SOYBN (Q96450) 14-3-3-like protein ... 141 1e-34
TC15496 homologue to UP|Q9LKL0 (Q9LKL0) 14-3-3 protein, partial ... 127 2e-30
TC15155 similar to UP|Q93XW1 (Q93XW1) 14-3-3 protein, partial (53%) 122 9e-29
TC15484 similar to GB|AAA96253.1|487791|ATU09376 GF14omega isofo... 104 2e-23
TC9741 homologue to GB|AAA96253.1|487791|ATU09376 GF14omega isof... 96 5e-21
AU251763 80 3e-16
AV420257 67 3e-12
TC11030 similar to UP|Q9M5K7 (Q9M5K7) 14-3-3-like protein, parti... 66 8e-12
TC12610 UP|Q944P2 (Q944P2) 14-3-3 protein, partial (21%) 48 2e-06
AU088886 45 1e-05
TC11586 similar to UP|O49152 (O49152) 14-3-3 protein homolog, pa... 37 0.005
TC7842 similar to UP|Q9XEW4 (Q9XEW4) 14-3-3 protein, partial (15%) 30 0.36
TC17939 weakly similar to UP|Q945P2 (Q945P2) AT5g49210/K21P3_8, ... 28 2.3
AU252170 27 4.0
>TC14325 homologue to UP|Q9FXL2 (Q9FXL2) Vf14-3-3c protein, partial (94%)
Length = 1080
Score = 219 bits (558), Expect(2) = 6e-81
Identities = 119/196 (60%), Positives = 147/196 (74%), Gaps = 8/196 (4%)
Frame = +2
Query: 9 SHNHTLTLDRKTK-----QSNKREKRIMGFTVAERLRREEYVFHAKLAQQAERYEEMVSF 63
SH T T++ +S+K++ + V E L RE+YV+ AKLA+QAERYEEMV F
Sbjct: 17 SHTFTHTVNNNNNNNHNLKSHKQKTMAVAGGVPENLSREQYVYLAKLAEQAERYEEMVEF 196
Query: 64 MQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAALRILS---KEEEGRKNEDDHFVHV 120
MQK+VVG TPA+ELS+EE NLLSVAYKN LRAA RI+S ++EE RKN DDH V V
Sbjct: 197 MQKLVVGSTPAAELSVEERNLLSVAYKNVIGSLRAAWRIVSSIEQKEESRKN-DDHVVLV 373
Query: 121 KKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVVYYQMKGDYQRYMAEFKIGDDKK 180
K Y+SKVE EL N+C SIL LLDS LIPSASSSE +V YY+MKGDY RY+AEF++GD +K
Sbjct: 374 KDYRSKVEVELSNICASILNLLDSNLIPSASSSESKVFYYKMKGDYHRYLAEFRVGDQRK 553
Query: 181 SAVEDIILSYKAAQDI 196
++ ED +LSYKAAQD+
Sbjct: 554 ASAEDTMLSYKAAQDL 601
Score = 98.2 bits (243), Expect(2) = 6e-81
Identities = 51/77 (66%), Positives = 61/77 (78%), Gaps = 1/77 (1%)
Frame = +1
Query: 196 IAAADLRSSHPIRLGLALNFSVFYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDS 254
+AA DL +HPIRLGLALNFSVFYYEILN+ D+ MA+QA +EA EL LG+E YKDS
Sbjct: 598 LAATDLPPTHPIRLGLALNFSVFYYEILNQSDKACAMAKQAFEEAIAELDTLGEESYKDS 777
Query: 255 TVRMQLLRNNIILWTFD 271
T+ MQLLR+N+ LWT D
Sbjct: 778 TLIMQLLRDNLTLWTSD 828
>TC7839 homologue to UP|Q93XW1 (Q93XW1) 14-3-3 protein, complete
Length = 1284
Score = 259 bits (661), Expect = 5e-70
Identities = 136/234 (58%), Positives = 175/234 (74%), Gaps = 3/234 (1%)
Frame = +3
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REEYV+ AKLA+QAERYEEMV FM+K+ EL++EE NLLSVAYKN RA+
Sbjct: 126 REEYVYMAKLAEQAERYEEMVEFMEKVSAA-ADTEELTVEERNLLSVAYKNVIGARRASW 302
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RI+S +++E + +DH ++ Y+SK+ESEL N+C IL+LLDS+LIPSASS + +V
Sbjct: 303 RIISSIEQKEESRGYEDHVSVIRDYRSKIESELSNICDGILKLLDSRLIPSASSGDSKVF 482
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
Y +MKGDY RY+AEFK G ++K A E + +YK+AQDIA ++L +HPIRLGLALNFSVF
Sbjct: 483 YLKMKGDYHRYLAEFKTGAERKEAAESTLAAYKSAQDIANSELPPTHPIRLGLALNFSVF 662
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEILN D ++A+QA DEA EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 663 YYEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSD 824
>TC7838 homologue to UP|Q93XW1 (Q93XW1) 14-3-3 protein, complete
Length = 1202
Score = 256 bits (654), Expect = 3e-69
Identities = 137/235 (58%), Positives = 175/235 (74%), Gaps = 4/235 (1%)
Frame = +3
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE+V+ AKLA+QAERYEEMV FM+K+ EL++EE NLLSVAYKN RA+
Sbjct: 156 REEFVYLAKLAEQAERYEEMVEFMEKLSAA-VDGEELTVEERNLLSVAYKNVIGARRASW 332
Query: 101 RILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S ++EE R NED H ++ Y+SK+ESEL N+C IL+LLDS+LIP+A+S + +V
Sbjct: 333 RIISSIEQKEESRGNED-HVSIIRDYRSKIESELSNICDGILKLLDSRLIPAAASGDSKV 509
Query: 158 VYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSV 217
Y +MKGDY RY+AEFK G ++K A E + +YK+AQDIA ++L +HPIRLGLALNFSV
Sbjct: 510 FYLKMKGDYHRYLAEFKTGTERKDAAESTLAAYKSAQDIANSELPPTHPIRLGLALNFSV 689
Query: 218 FYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
FYYEILN D +A+QA DEA EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 690 FYYEILNSPDRACSLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSD 854
>TC15349 homologue to UP|143C_SOYBN (Q96452) 14-3-3-like protein C (SGF14C),
complete
Length = 1149
Score = 225 bits (574), Expect = 6e-60
Identities = 122/241 (50%), Positives = 164/241 (67%), Gaps = 3/241 (1%)
Frame = +1
Query: 34 TVAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNAT 93
++A R+ +V+ AKLA+QAERYEEMV M+ + EL++EE NLLSV YKN
Sbjct: 67 SMASTKERDNFVYIAKLAEQAERYEEMVEAMKNVAKLNV---ELTVEERNLLSVGYKNVV 237
Query: 94 EPLRAALRILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSAS 151
RA+ RILS +++E K D +++Y+ KVESEL N+C I+ ++D LIPS+S
Sbjct: 238 GARRASWRILSSIEQKEEAKGNDVSVKRIREYRQKVESELSNICSDIMIVIDEHLIPSSS 417
Query: 152 SSEIRVVYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGL 211
+ E V +Y+MKGDY RY+AEFK GDD+K A + + +Y+ A A +L +HPIRLGL
Sbjct: 418 AGEPSVFFYKMKGDYYRYLAEFKSGDDRKEAADQSMKAYQLASTTAETELPPTHPIRLGL 597
Query: 212 ALNFSVFYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTF 270
ALNFSVFYYEILN + +A+QA DEA +EL L +E YKDST+ MQLLR+N+ LWT
Sbjct: 598 ALNFSVFYYEILNSPERACHLAKQAFDEAISELDTLSEESYKDSTLIMQLLRDNLTLWTS 777
Query: 271 D 271
D
Sbjct: 778 D 780
>TC9505 similar to PIR|F86391|F86391 T1K7.15 protein - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (62%)
Length = 943
Score = 172 bits (435), Expect = 8e-44
Identities = 86/153 (56%), Positives = 109/153 (71%), Gaps = 1/153 (0%)
Frame = +1
Query: 120 VKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVVYYQMKGDYQRYMAEFKIGDDK 179
+K Y+ KVE EL +CG IL ++D LIPS++S+E V YY+MKGDY RY+AEFK ++
Sbjct: 25 IKNYRQKVEEELSKICGDILTIIDQHLIPSSASAEASVFYYKMKGDYYRYLAEFKTDQER 204
Query: 180 KSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVFYYEILNRFDEGLDMARQALDE 239
K A E + Y+AA A DL S+HPIRLGLALNFSVFYYEI+N + +A+QA DE
Sbjct: 205 KEAAEQSLKGYEAASATANTDLPSTHPIRLGLALNFSVFYYEIMNSPERACHLAKQAFDE 384
Query: 240 ARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
A EL L +E YKDST+ MQLLR+N+ LWT D
Sbjct: 385 AIAELDTLSEESYKDSTLIMQLLRDNLTLWTSD 483
>TC16771 homologue to UP|O49152 (O49152) 14-3-3 protein homolog, partial
(61%)
Length = 550
Score = 142 bits (359), Expect = 5e-35
Identities = 75/155 (48%), Positives = 107/155 (68%), Gaps = 2/155 (1%)
Frame = +1
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
RE +V+ AKLA+QAERYEEMV M+K+ EL++EE NLLSV YKN RA+
Sbjct: 88 RETFVYVAKLAEQAERYEEMVDSMKKVA---NLDVELTVEERNLLSVGYKNVIGARRASW 258
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RILS +++E K D + +K+Y+ KVESEL ++C ++ ++D LIPSA++ E V
Sbjct: 259 RILSSIEQKEETKGNDVNAKRIKEYRQKVESELADICNDVMRVIDEHLIPSATAGESTVF 438
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAA 193
YY+MKGDY RY+AEFK G++KK A + + +Y++A
Sbjct: 439 YYKMKGDYYRYLAEFKSGNEKKEAADQSMKAYESA 543
>TC12158 homologue to UP|143A_SOYBN (Q96450) 14-3-3-like protein A (SGF14A),
partial (54%)
Length = 562
Score = 141 bits (356), Expect = 1e-34
Identities = 77/137 (56%), Positives = 99/137 (72%), Gaps = 3/137 (2%)
Frame = +2
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE V+ AKLA+QAERYEEMV FM+K+ A EL++EE NLLSVAYKN RA+
Sbjct: 155 REENVYLAKLAEQAERYEEMVEFMEKVAKS-VDAEELTVEERNLLSVAYKNVIGARRASW 331
Query: 101 RILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S ++EE R NED H +K+Y+ K+E+EL +C IL LL+S LIPSA++ E +V
Sbjct: 332 RIISSIEQKEESRGNED-HVSVIKEYRGKIEAELSKICDGILNLLESNLIPSAAAPESKV 508
Query: 158 VYYQMKGDYQRYMAEFK 174
Y +MKGDY RY+AEFK
Sbjct: 509 FYLKMKGDYHRYLAEFK 559
>TC15496 homologue to UP|Q9LKL0 (Q9LKL0) 14-3-3 protein, partial (48%)
Length = 677
Score = 127 bits (320), Expect = 2e-30
Identities = 65/103 (63%), Positives = 81/103 (78%), Gaps = 1/103 (0%)
Frame = +3
Query: 170 MAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVFYYEILNRFDEG 229
+AEFK G ++K A E +L+YK+AQDIA A+L +HPIRLGLALNFSVFYYEILN D
Sbjct: 3 LAEFKTGAERKEAAESTLLAYKSAQDIALAELAPTHPIRLGLALNFSVFYYEILNSPDRA 182
Query: 230 LDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
++A+QA DEA +EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 183 CNLAKQAFDEAISELDTLGEESYKDSTLIMQLLRDNLTLWTSD 311
>TC15155 similar to UP|Q93XW1 (Q93XW1) 14-3-3 protein, partial (53%)
Length = 548
Score = 122 bits (305), Expect = 9e-29
Identities = 64/132 (48%), Positives = 91/132 (68%), Gaps = 3/132 (2%)
Frame = +3
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGY-TPASELSLEEMNLLSVAYKNATEPLRAA 99
RE+Y++ AKLA++AE Y+EMV FM+K+ EL++EE N LSVAYKN RA+
Sbjct: 153 REKYLYMAKLAEEAEAYDEMVEFMEKVYAAADNEDQELTVEERNHLSVAYKNVMGTKRAS 332
Query: 100 LRILSKEE--EGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S +E E +DH ++++++ VES L N+C +L LLDS+LIPSA+S+E +V
Sbjct: 333 WRIISSKEAKEESLGNEDHLDNIREFRYMVESHLCNICDDVLNLLDSRLIPSATSAESKV 512
Query: 158 VYYQMKGDYQRY 169
Y +MKGDY RY
Sbjct: 513 FYLKMKGDYYRY 548
>TC15484 similar to GB|AAA96253.1|487791|ATU09376 GF14omega isoform
{Arabidopsis thaliana;} , partial (37%)
Length = 655
Score = 104 bits (259), Expect = 2e-23
Identities = 53/84 (63%), Positives = 67/84 (79%), Gaps = 1/84 (1%)
Frame = +3
Query: 187 ILSYKAAQDIAAADLRSSHPIRLGLALNFSVFYYEILNRFDEGLDMARQALDEARNEL-K 245
+++YK+A+DIA A+L +HPIRLGLALNFSVFYYEILN D +A+ A DEA +EL
Sbjct: 3 LVAYKSAEDIANAELAPTHPIRLGLALNFSVFYYEILNSPDRACKIAKDAFDEAISELDT 182
Query: 246 LGDEYYKDSTVRMQLLRNNIILWT 269
LG+E YKDST+ MQLLR+N+ LWT
Sbjct: 183 LGEESYKDSTLIMQLLRDNLTLWT 254
>TC9741 homologue to GB|AAA96253.1|487791|ATU09376 GF14omega isoform
{Arabidopsis thaliana;} , partial (32%)
Length = 507
Score = 96.3 bits (238), Expect = 5e-21
Identities = 50/77 (64%), Positives = 60/77 (76%), Gaps = 1/77 (1%)
Frame = +3
Query: 194 QDIAAADLRSSHPIRLGLALNFSVFYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYK 252
+DIA A+L +HPIRLGLALNFSVFYYEILN D +A+ A DEA +EL LG+E YK
Sbjct: 3 EDIANAELAPTHPIRLGLALNFSVFYYEILNSPDRACKIAKDAFDEAISELDTLGEESYK 182
Query: 253 DSTVRMQLLRNNIILWT 269
DST+ MQLLR+N+ LWT
Sbjct: 183 DSTLIMQLLRDNLTLWT 233
>AU251763
Length = 340
Score = 80.5 bits (197), Expect = 3e-16
Identities = 49/115 (42%), Positives = 68/115 (58%), Gaps = 2/115 (1%)
Frame = +3
Query: 34 TVAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNAT 93
T+A RE V+ AKL++QAERYEEMV M+ + EL++EE NLLSV YKN
Sbjct: 3 TMAAEKERETQVYMAKLSEQAERYEEMVECMKAVA---KLDLELTVEERNLLSVGYKNVI 173
Query: 94 EPLRAALRILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKL 146
RA+ RI+S +++E K + + +K Y KVE EL +C IL ++D L
Sbjct: 174 GARRASWRIMSSIEQKEESKGNESNVKLIKGYCHKVEEELSKICIDILTIIDQHL 338
>AV420257
Length = 256
Score = 67.0 bits (162), Expect = 3e-12
Identities = 36/85 (42%), Positives = 51/85 (59%), Gaps = 2/85 (2%)
Frame = +1
Query: 79 LEEMNLLSVAYKNATEPLRAALRILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCG 136
+EE NL SV YKN RA+ RILS +++E K + ++ Y+ KVE EL N+C
Sbjct: 1 VEERNLFSVGYKNVVGSRRASWRILSSIEQKEESKGNELSVKRIRDYRHKVELELSNICS 180
Query: 137 SILELLDSKLIPSASSSEIRVVYYQ 161
I+ +LD LIPS + +E V YY+
Sbjct: 181 DIMIILDEHLIPSTNVAESTVFYYK 255
>TC11030 similar to UP|Q9M5K7 (Q9M5K7) 14-3-3-like protein, partial (32%)
Length = 564
Score = 65.9 bits (159), Expect = 8e-12
Identities = 34/61 (55%), Positives = 45/61 (73%), Gaps = 1/61 (1%)
Frame = +2
Query: 197 AAADLRSSHPIRLGLALNFSVFYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDST 255
A ++LR +HPIRLGLALNFSVFYYEI+N +A+ A D+A +EL L ++ YKDST
Sbjct: 2 AESELRRTHPIRLGLALNFSVFYYEIMNSPARACRLAKPAFDDAVSELDTLNEDSYKDST 181
Query: 256 V 256
+
Sbjct: 182 L 184
>TC12610 UP|Q944P2 (Q944P2) 14-3-3 protein, partial (21%)
Length = 534
Score = 47.8 bits (112), Expect = 2e-06
Identities = 25/41 (60%), Positives = 31/41 (74%), Gaps = 1/41 (2%)
Frame = +2
Query: 232 MARQALDEARNELK-LGDEYYKDSTVRMQLLRNNIILWTFD 271
+A+QA DEA EL L +E YKDST+ MQLLR+N+ LWT D
Sbjct: 23 LAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSD 145
>AU088886
Length = 327
Score = 45.1 bits (105), Expect = 1e-05
Identities = 29/68 (42%), Positives = 38/68 (55%)
Frame = +3
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE+V+ AKLA+QAERYE + + +NLLSVAYKN RA+
Sbjct: 42 REEFVYLAKLAEQAERYEXDG*VHGEALRRRRRRRNSPSXSVNLLSVAYKNVIGARRASW 221
Query: 101 RILSKEEE 108
RI+S E+
Sbjct: 222 RIISSIEQ 245
>TC11586 similar to UP|O49152 (O49152) 14-3-3 protein homolog, partial (18%)
Length = 553
Score = 36.6 bits (83), Expect = 0.005
Identities = 16/23 (69%), Positives = 19/23 (82%)
Frame = +2
Query: 249 EYYKDSTVRMQLLRNNIILWTFD 271
E YKDST+ MQLLR+N+ LWT D
Sbjct: 2 ESYKDSTLIMQLLRDNLTLWTSD 70
>TC7842 similar to UP|Q9XEW4 (Q9XEW4) 14-3-3 protein, partial (15%)
Length = 403
Score = 30.4 bits (67), Expect = 0.36
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +3
Query: 252 KDSTVRMQLLRNNIILWTFD 271
KDST+ M LLR+ + LWTFD
Sbjct: 18 KDSTLIMPLLRDYLTLWTFD 77
>TC17939 weakly similar to UP|Q945P2 (Q945P2) AT5g49210/K21P3_8, partial
(61%)
Length = 999
Score = 27.7 bits (60), Expect = 2.3
Identities = 19/67 (28%), Positives = 32/67 (47%)
Frame = +2
Query: 48 AKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAALRILSKEE 107
AKLAQ Y + V ++K + EEM L+ + + E R ALR+ ++E
Sbjct: 302 AKLAQLRRDYAKQVKELRKEYI----------EEMELMRLEKQRKDEARREALRVATEER 451
Query: 108 EGRKNED 114
+ K ++
Sbjct: 452 KRLKAQE 472
>AU252170
Length = 350
Score = 26.9 bits (58), Expect = 4.0
Identities = 15/33 (45%), Positives = 20/33 (60%), Gaps = 2/33 (6%)
Frame = -3
Query: 32 GFTVAERLRREEYVFHAKLAQQAERY--EEMVS 62
GFT+A R RRE+ F ++ Q RY +EM S
Sbjct: 144 GFTIAARRRREDR*FSGEIELQFRRYSRDEMKS 46
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,046,355
Number of Sequences: 28460
Number of extensions: 41304
Number of successful extensions: 234
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 209
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 210
length of query: 286
length of database: 4,897,600
effective HSP length: 89
effective length of query: 197
effective length of database: 2,364,660
effective search space: 465838020
effective search space used: 465838020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC141414.9


