

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141114.10 + phase: 0
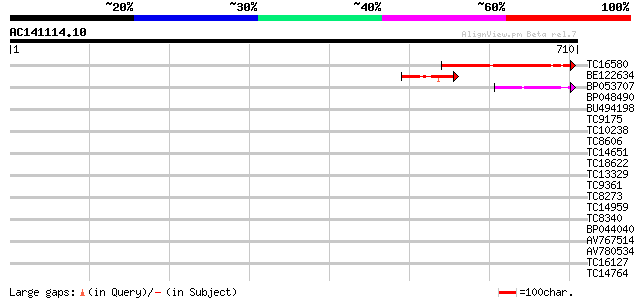
(710 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16580 147 4e-36
BE122634 67 7e-12
BP053707 58 5e-09
BP048490 34 0.070
BU494198 33 0.16
TC9175 weakly similar to UP|INAD_DROME (Q24008) Inactivation-no-... 31 0.60
TC10238 31 0.60
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 30 1.7
TC14651 weakly similar to UP|Q42779 (Q42779) Alpha-carboxyltrans... 30 1.7
TC18622 weakly similar to UP|AAR06917 (AAR06917) UDP-glycosyltra... 30 1.7
TC13329 similar to GB|AAC42247.2|20197365|AC005395 expressed pro... 29 3.0
TC9361 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (68%) 29 3.0
TC8273 UP|BAD11134 (BAD11134) Glutamate-rich protein, complete 29 3.0
TC14959 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5... 28 3.9
TC8340 similar to UP|SEC8_ARATH (Q93YU5) Probable exocyst comple... 28 5.0
BP044040 28 6.6
AV767514 27 8.6
AV780534 27 8.6
TC16127 similar to GB|AAF87136.1|9369388|AC002423 T23E23.3 {Arab... 27 8.6
TC14764 similar to UP|O22198 (O22198) 4-alpha-glucanotransferase... 27 8.6
>TC16580
Length = 683
Score = 147 bits (372), Expect = 4e-36
Identities = 79/168 (47%), Positives = 109/168 (64%)
Frame = +2
Query: 541 SDLLMLPKDMLMDRQVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALNGETIV 600
SDLLMLPKDML+ + +EVCP + S I ++L NF PDEFCPD +P V EAL+ + +
Sbjct: 2 SDLLMLPKDMLLSESIRKEVCPMFNASQIKKILDNFVPDEFCPDPIPTDVFEALDSKDDL 181
Query: 601 ERRMSAESIRSFPYSAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEEL 660
E +S+ +FP AAP+VY PP + +A + G + L R+ S+V R+ YTSD+EL
Sbjct: 182 ED--GKDSVNNFPCIAAPIVYSPPPATTIASITGDIGSESQLRRSQSSVVRKSYTSDDEL 355
Query: 661 EELDSPLSSIIDKVPSSPTVATNGNGNHEEQGSQTTTNARYQLLREVW 708
+EL+SPLSSI+ SP V+T N +E S+T + RY+LLR VW
Sbjct: 356 DELNSPLSSILFSGSPSP-VSTKPNWKKKE--SRTESAVRYELLRNVW 490
>BE122634
Length = 301
Score = 67.4 bits (163), Expect = 7e-12
Identities = 37/75 (49%), Positives = 50/75 (66%), Gaps = 3/75 (4%)
Frame = +1
Query: 491 NSVGNWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFA---LLNDLSDLLMLP 547
+S+G+WSR L+D+FGID D E+ ++END GP + SF LLN LSDL+MLP
Sbjct: 88 DSIGDWSRWLSDLFGIDDSDSHED---SNEND----GPKYESSFKPFPLLNSLSDLMMLP 246
Query: 548 KDMLMDRQVSQEVCP 562
+ML D + +EVCP
Sbjct: 247 SEMLADGSMRKEVCP 291
>BP053707
Length = 476
Score = 58.2 bits (139), Expect = 5e-09
Identities = 42/103 (40%), Positives = 54/103 (51%), Gaps = 2/103 (1%)
Frame = -2
Query: 608 SIRSFPYSAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEELDSPL 667
SI SFP SA Y PP + +V + E G L R+ S V ++ YTSD+EL+ELDSPL
Sbjct: 457 SITSFPCSAGSTFYEPPPARSVVXMLKEVGTPNSL-RSGSFVLQKLYTSDDELDELDSPL 281
Query: 668 SSI--IDKVPSSPTVATNGNGNHEEQGSQTTTNARYQLLREVW 708
S++ D S G G + RY+LLREVW
Sbjct: 280 SALGMDDSSLGSMKKFAVGKGGRKV--------VRYELLREVW 176
>BP048490
Length = 452
Score = 34.3 bits (77), Expect = 0.070
Identities = 27/94 (28%), Positives = 45/94 (47%), Gaps = 4/94 (4%)
Frame = +3
Query: 59 KVRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKV 118
K ++ ES+ S E +D K + S KKS +S+ ++ ++ + V
Sbjct: 12 KSESSDDESSESSDEENDSKP-ATTTVAKPSAVKKSASSDSDDEDSSDSDSEVKKDKMDV 188
Query: 119 PAKVSSESSEGVDEK----PVIEVKEIEILDGSS 148
SSE S+ DEK P +VK++E++D SS
Sbjct: 189 DESDSSEDSDEEDEKSTKTPQKKVKDVEMVDASS 290
>BU494198
Length = 510
Score = 33.1 bits (74), Expect = 0.16
Identities = 27/107 (25%), Positives = 48/107 (44%), Gaps = 6/107 (5%)
Frame = +3
Query: 57 KNKVRANSLESNRGSRERSDRKTNKLQS------KVSGSNQKKSINSNKGPSRVTNKNTS 110
+ K ++ + +S++ SR + D T K K S Q+ S + ++ S
Sbjct: 12 EQKSKSKNTDSSKISRSKDDVSTPKSAKSKHETPKTGKSKQETSKIAAASKAKSPKSGKS 191
Query: 111 TNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSE 157
P KV + VS +S++ DE +E+E G ++ + VGSE
Sbjct: 192 NTNGPGKVKS-VSLKSTDSEDETSDDSTREVEDTKGKTSTSSKVGSE 329
>TC9175 weakly similar to UP|INAD_DROME (Q24008)
Inactivation-no-after-potential D protein, partial (5%)
Length = 692
Score = 31.2 bits (69), Expect = 0.60
Identities = 24/91 (26%), Positives = 36/91 (39%), Gaps = 1/91 (1%)
Frame = +2
Query: 107 KNTSTNTKPVKVPAKVSSESSEGVDE-KPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVN 165
+ T K V P + EG +E KP E + E A+ E+ E +
Sbjct: 23 RRTKKQAKIVPQPEPEPEKKEEGKEEEKPAAEEGKPEEKKEEEKAAEEAKKEEGGEEKND 202
Query: 166 AEENGEGEDGAAMKSKIKEMESRIENLEEEL 196
+E GE+G K + + N+EEEL
Sbjct: 203 NKEEKGGEEGKEEGKKEENNNGAVVNMEEEL 295
>TC10238
Length = 942
Score = 31.2 bits (69), Expect = 0.60
Identities = 17/37 (45%), Positives = 27/37 (72%)
Frame = -1
Query: 63 NSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNK 99
+S +S G + ++++T ++KVS +NQKKSINSNK
Sbjct: 768 SSSKSIIGDPKLNEKET---ETKVSNNNQKKSINSNK 667
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 29.6 bits (65), Expect = 1.7
Identities = 23/86 (26%), Positives = 38/86 (43%), Gaps = 3/86 (3%)
Frame = -2
Query: 92 KKSINSNKGPSRVTNKNTSTNTK--PVKVPAKVSSESSEGVDEKPVI-EVKEIEILDGSS 148
++ ++S G N N +N+K P P + G DE+ E +E + D
Sbjct: 551 EEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDEEDEDGEDQEDDDDDDED 372
Query: 149 NGAQSVGSEDEIHEIVNAEENGEGED 174
+ + G EDE E V E+N + E+
Sbjct: 371 DDEEDDGGEDEEEEGVEEEDNEDEEE 294
>TC14651 weakly similar to UP|Q42779 (Q42779) Alpha-carboxyltransferase
aCT-1 precursor , partial (22%)
Length = 1317
Score = 29.6 bits (65), Expect = 1.7
Identities = 33/140 (23%), Positives = 55/140 (38%), Gaps = 8/140 (5%)
Frame = +1
Query: 45 EDVKVEKASKDPKNKVRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRV 104
+++++ +S D K+K+ L +G R+ S++ N + GP R
Sbjct: 523 KEIEISASSSDVKSKIEQLKLGGCQGWRDT*--------SRIKEKNYFFDATN*AGPCR- 675
Query: 105 TNKNTSTNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIV 164
+ ++S K + +KVSS+ E +DG G S + DE+ E V
Sbjct: 676 GHHSSSIKEKYDNLVSKVSSKPEE---------------VDGGLRGDSSTTTNDELREKV 810
Query: 165 NAEEN--------GEGEDGA 176
A G EDGA
Sbjct: 811 GANRTIS*NFCSYGASEDGA 870
>TC18622 weakly similar to UP|AAR06917 (AAR06917) UDP-glycosyltransferase
73E1, partial (14%)
Length = 424
Score = 29.6 bits (65), Expect = 1.7
Identities = 14/57 (24%), Positives = 30/57 (52%), Gaps = 2/57 (3%)
Frame = +3
Query: 131 DEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNA--EENGEGEDGAAMKSKIKEM 185
+EK +++V + + G G ++ ++++ + + E+ GEGE K+KEM
Sbjct: 57 NEKLIVQVLKTGVRIGVEGGGDTIVKKEDVKKAIEQLMEQGGEGEQRRNRAKKLKEM 227
>TC13329 similar to GB|AAC42247.2|20197365|AC005395 expressed protein
{Arabidopsis thaliana;} , partial (9%)
Length = 576
Score = 28.9 bits (63), Expect = 3.0
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 4/81 (4%)
Frame = -1
Query: 155 GSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEELREVAALEVSLYSI----V 210
GSE H+ VN E+ E + +KSKIKE++S + + + L V LYS +
Sbjct: 576 GSEKITHQ-VNRED*---EYSSNLKSKIKEIKSVTN*INAQHKRNQVLRVFLYSFEDFNL 409
Query: 211 PEHGSSAHKVHTPARRLSRLY 231
P S++ H R S +Y
Sbjct: 408 PSFSHSSYSKHALLIRPSFIY 346
>TC9361 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (68%)
Length = 781
Score = 28.9 bits (63), Expect = 3.0
Identities = 17/52 (32%), Positives = 27/52 (51%)
Frame = +2
Query: 611 SFPYSAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEE 662
S P +A P V P++ ++ G R +S+ QRR +SD+E+EE
Sbjct: 422 SAPTTATPTVMTAPTTTKPLDEKPSRG------RILSSEQRRSSSSDDEIEE 559
>TC8273 UP|BAD11134 (BAD11134) Glutamate-rich protein, complete
Length = 923
Score = 28.9 bits (63), Expect = 3.0
Identities = 25/130 (19%), Positives = 49/130 (37%)
Frame = +1
Query: 50 EKASKDPKNKVRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNT 109
E + P ++V A + + + K+++ + + + + P+ V K
Sbjct: 142 ETTIEQPASEVPAPEPTIEQEHPKEETTEEPKVEAVAETTTEAPAAPETEAPAEVETKEV 321
Query: 110 STNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEEN 169
+ TKP + E V E+P++E E + ++ A V E E AE
Sbjct: 322 TEETKPEAEKTEEPEVKPEEVKEEPIVE----ETKETAATEAAPVEEEKSSEESKAAETT 489
Query: 170 GEGEDGAAMK 179
E A++
Sbjct: 490 VEATTEVAIE 519
>TC14959 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;}, partial (16%)
Length = 1015
Score = 28.5 bits (62), Expect = 3.9
Identities = 20/77 (25%), Positives = 35/77 (44%), Gaps = 5/77 (6%)
Frame = +2
Query: 141 IEILDGSSNGAQSVGSEDE-----IHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEE 195
+E+L G + S+G E + +V E E D M R +N+EEE
Sbjct: 188 LELLTGKAPNQASLGEEGIDLPRWVQSVVREEWTAEVFDAELM---------RFQNIEEE 340
Query: 196 LREVAALEVSLYSIVPE 212
+ ++ + ++ SIVP+
Sbjct: 341 MVQLLQIAMACVSIVPD 391
>TC8340 similar to UP|SEC8_ARATH (Q93YU5) Probable exocyst complex component
Sec8, partial (21%)
Length = 1198
Score = 28.1 bits (61), Expect = 5.0
Identities = 17/41 (41%), Positives = 23/41 (55%), Gaps = 2/41 (4%)
Frame = -3
Query: 145 DGSSNGAQSVGSEDEI-HEIVNAE-ENGEGEDGAAMKSKIK 183
D S AQ S ++ H I+N + EGE+G A+K KIK
Sbjct: 1031 DRESEKAQKESSSNQN*HHIINYNLQTNEGENGTAIKGKIK 909
>BP044040
Length = 491
Score = 27.7 bits (60), Expect = 6.6
Identities = 30/106 (28%), Positives = 46/106 (43%), Gaps = 7/106 (6%)
Frame = +3
Query: 55 DPKNKVRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKS-----INSNKGPS-RVT-NK 107
D K+KV+ + N S K K Q +V GS+ S IN N+ P+ R+T +
Sbjct: 57 DMKDKVKGFMKKVNNPFTSSSSGKF-KGQGRVLGSSSSSSSSSAPINPNRNPTPRLTPTQ 233
Query: 108 NTSTNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQS 153
N + N+KP S + + P + L SSN +Q+
Sbjct: 234 NPNPNSKPRPASTTNHEPSPQKTNRNPGDGFDPFDSLVTSSNRSQN 371
>AV767514
Length = 597
Score = 27.3 bits (59), Expect = 8.6
Identities = 12/28 (42%), Positives = 18/28 (63%)
Frame = -2
Query: 340 GTFTLALERVESWIFSRLVESVWWQALT 367
G+FTL LE+ +W+ L +V W +LT
Sbjct: 104 GSFTLFLEKSPTWLGDFLAVAVSWLSLT 21
>AV780534
Length = 536
Score = 27.3 bits (59), Expect = 8.6
Identities = 23/93 (24%), Positives = 44/93 (46%), Gaps = 8/93 (8%)
Frame = +1
Query: 50 EKASKDPKNKVRANSLESNRGSRERSDRK---TNKLQSKVSGSNQK-----KSINSNKGP 101
EK P ++++ ++ +S R+RS R +N + ++ S K KSIN
Sbjct: 70 EKNDYQPVSEIKDSNDKSGWSFRKRSARHRVLSNTVITETPSSANKETSECKSINFQPLE 249
Query: 102 SRVTNKNTSTNTKPVKVPAKVSSESSEGVDEKP 134
+ T++ TS N +P + + ++ D+KP
Sbjct: 250 PKETSECTSINVQPQEPNVAEKNNTTNFSDDKP 348
>TC16127 similar to GB|AAF87136.1|9369388|AC002423 T23E23.3 {Arabidopsis
thaliana;} , partial (6%)
Length = 442
Score = 27.3 bits (59), Expect = 8.6
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = +1
Query: 563 SISLSLIIRVLCNFTP 578
S+ LSL +RV CNFTP
Sbjct: 7 SLQLSLSLRVCCNFTP 54
>TC14764 similar to UP|O22198 (O22198) 4-alpha-glucanotransferase, partial
(10%)
Length = 608
Score = 27.3 bits (59), Expect = 8.6
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 5/52 (9%)
Frame = +3
Query: 610 RSFPYSAAPVVYMPPSSVNVAE-----KVAEAGGKCHLTRNVSAVQRRGYTS 656
RSFP+ + V P S+ +VAE K+A A K L + VQ R + +
Sbjct: 207 RSFPHEDSQVEASPVSASSVAEAVSDKKIAGAADKIRLPSESNGVQPRDHVA 362
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.128 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,636,795
Number of Sequences: 28460
Number of extensions: 134477
Number of successful extensions: 631
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 620
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 624
length of query: 710
length of database: 4,897,600
effective HSP length: 97
effective length of query: 613
effective length of database: 2,136,980
effective search space: 1309968740
effective search space used: 1309968740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141114.10


