

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141111.7 - phase: 0 /pseudo
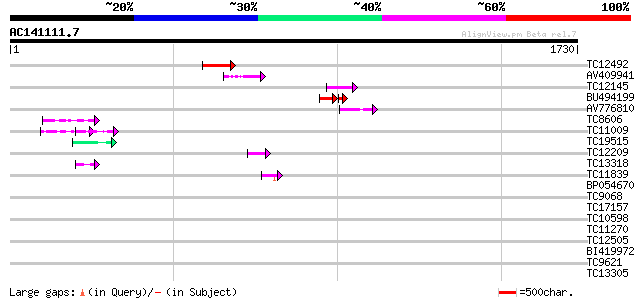
(1730 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, parti... 103 3e-22
AV409941 71 2e-12
TC12145 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment)... 71 2e-12
BU494199 47 2e-12
AV776810 64 2e-10
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 55 9e-08
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 54 3e-07
TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, pa... 48 1e-05
TC12209 similar to UP|O60177 (O60177) DEAD box helicase, partial... 44 2e-04
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 44 3e-04
TC11839 similar to UP|STH1_YEAST (P32597) Nuclear protein STH1/N... 43 5e-04
BP054670 42 0.001
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 41 0.002
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 40 0.002
TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526 At2g01... 40 0.004
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 39 0.005
TC12505 similar to UP|BAD00698 (BAD00698) Sericin 1A, partial (5%) 39 0.009
BI419972 38 0.012
TC9621 similar to UP|Q9SIW2 (Q9SIW2) At2g16390 protein, partial ... 38 0.016
TC13305 weakly similar to UP|O29528 (O29528) Cobalt transport pr... 37 0.026
>TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, partial (7%)
Length = 922
Score = 103 bits (256), Expect = 3e-22
Identities = 45/101 (44%), Positives = 67/101 (65%)
Frame = +1
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
K+ EQP L+GG+LR YQ+EGL ++++ + N+ N +LADEMGLGKT+Q++S++ L +
Sbjct: 619 KVTEQPSILQGGELRSYQIEGLQWMLSLFNNNLNGILADEMGLGKTIQTISLIAHLMEYK 798
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSR 687
+ GP L+V P + L NW EF W P + I+Y G R
Sbjct: 799 GVTGPHLIVAPKAVLPNWMNEFSTWAPSIKTILYDGRMDER 921
>AV409941
Length = 429
Score = 70.9 bits (172), Expect = 2e-12
Identities = 43/131 (32%), Positives = 70/131 (52%)
Frame = +1
Query: 651 PFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNAL 710
P L++ P S + NW EF KW + +V +Y G ++R++ N G +I L
Sbjct: 67 PVLIICPSSVIHNWEDEFSKW-SNFSVSIYHG--ANRDLVYDKLQAN----GVEI----L 213
Query: 711 LTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNS 770
+T+++ I WN +++DEAHRLKN ++LY A E T + +TGT +QN
Sbjct: 214 ITSFDSYRIHGRSFLDINWNVVIIDEAHRLKNDRSKLYKACLEIKTLRRYGLTGTVMQNK 393
Query: 771 VEELWALLHFL 781
+ EL+ L ++
Sbjct: 394 IMELFNLFDWV 426
>TC12145 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment), partial
(76%)
Length = 664
Score = 70.9 bits (172), Expect = 2e-12
Identities = 37/95 (38%), Positives = 55/95 (56%)
Frame = +2
Query: 966 AMDHFNAPGSDDFCFLLSTRAGGLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQRE 1025
A+ FN F FLLS +A G+ +NL A V + + WNP + QA R HRIGQ +
Sbjct: 11 AIKRFNDDPDCKF-FLLSLKAAGVALNLTVASHVFLMEPWWNPDAEQQAQDRIHRIGQNK 187
Query: 1026 VVNIYRFVTSKSVEEDILERAKKKMVLDHLVIQKL 1060
+ + +F+T +VEE ILE +KK + +I++L
Sbjct: 188 PIRVVKFITENTVEERILELQQKKEAVSEGLIRRL 292
>BU494199
Length = 570
Score = 47.0 bits (110), Expect(2) = 2e-12
Identities = 18/29 (62%), Positives = 24/29 (82%)
Frame = +2
Query: 1003 DSDWNPQNDLQAMSRAHRIGQREVVNIYR 1031
+ DWNP D QAM RAHR+GQ++VVN++R
Sbjct: 431 EHDWNPMRDHQAMDRAHRLGQKKVVNVHR 517
Score = 43.5 bits (101), Expect(2) = 2e-12
Identities = 22/55 (40%), Positives = 35/55 (63%)
Frame = +1
Query: 946 LRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATADTVI 1000
++ + RLDGS + E R + + FN+ + D LL+T GGLG+NL +ADT++
Sbjct: 262 MKNVTYLRLDGSVEPEKRFEIVKAFNSDPTIDV-LLLTTHVGGLGLNLTSADTLV 423
>AV776810
Length = 360
Score = 63.9 bits (154), Expect = 2e-10
Identities = 38/116 (32%), Positives = 60/116 (50%)
Frame = +2
Query: 1006 WNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVIQKLNAEGK 1065
WNP D QA R HRIGQ V+IYR ++ ++EE+IL++A +K LD LVIQ +
Sbjct: 2 WNPAMDQQAQDRCHRIGQTREVHIYRLISESTIEENILKKANQKRALDDLVIQSGGYNTE 181
Query: 1066 LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEILERAE 1121
KK D E+ + R + + +E+N + + + D++ L+ E
Sbjct: 182 FFKK--------LDPMEIFSGHRTLSIKNTPKEKNQNNGEVSVTNADVEAALKYVE 325
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 55.1 bits (131), Expect = 9e-08
Identities = 43/173 (24%), Positives = 72/173 (40%)
Frame = -2
Query: 101 SKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQD 160
+K + ++ED+ D E + G ED+DG G+G+ E++ S+D G
Sbjct: 668 TKGETKSQSEDKQHDAEGEDGNDDEGDEDDDGDGPFGEGE--------EDLSSEDGAGYG 513
Query: 161 GEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDE 220
+S + P G + P + E+D+D D E+D+ D+
Sbjct: 512 NNSNNKSNSKKA--PEGGPGAGAGPEENG-------------EDEEDEDGEDQEDDDDDD 378
Query: 221 DDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAK 273
+D D+ D + +D E E +E D+ DE+ D D + L KK K
Sbjct: 377 EDDDEED----------DGGEDEEEEGVEEEDNEDEEEDEEDEEALQPPKKRK 249
Score = 31.6 bits (70), Expect = 1.1
Identities = 31/124 (25%), Positives = 46/124 (37%)
Frame = -2
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVS 261
+D + D + D +DE DEDD D F EGE+ D S ED
Sbjct: 641 EDKQHDAEGEDGNDDEGDEDDDGDGPFG--------------EGEE----DLSSEDGAGY 516
Query: 262 DNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSE 321
N+ + K+ ++ + GP A + EDED +D D +
Sbjct: 515 GNNS---NNKSNSKKAPEGGPG-----------AGAGPEENGEDEEDEDGEDQEDDDDDD 378
Query: 322 SDED 325
D+D
Sbjct: 377 EDDD 366
Score = 30.4 bits (67), Expect = 2.5
Identities = 26/132 (19%), Positives = 55/132 (40%), Gaps = 1/132 (0%)
Frame = -2
Query: 33 EAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSSYLKDCQPMS-P 91
E + +S+ + DA G+ D + + S+ + G+ Y + S
Sbjct: 659 ETKSQSEDKQHDAEGEDGNDDEGDEDDDGDGPFGEGEEDLSSED--GAGYGNNSNNKSNS 486
Query: 92 QNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEM 151
+ E G + +G + EDE +++ + + ED+D + D G+ + + + EE
Sbjct: 485 KKAPEGGPGAGAGPEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDE--EEEGVEEED 312
Query: 152 LSDDSYGQDGEE 163
D+ ++ EE
Sbjct: 311 NEDEEEDEEDEE 276
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 53.5 bits (127), Expect = 3e-07
Identities = 50/170 (29%), Positives = 70/170 (40%), Gaps = 5/170 (2%)
Frame = +2
Query: 93 NGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSED---EDGQKDSGKGQRGDSDVPAE 149
+G E DD D DE +D+ + G G+ E+ E+G + G+G GD D
Sbjct: 20 SGDEDDDDDGEDDD---GDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPD---- 178
Query: 150 EMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDD 209
DD D +E+ E G T V R V AED++
Sbjct: 179 ---DDDDDDDDDDEEEEEEEDLG-------------TEYLVRRTVAA-------AEDEEA 289
Query: 210 DADYE--EDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDED 257
+D+E EDE D++D DD + S R K D +G D+ DD ED
Sbjct: 290 SSDFEPEEDEGDDNDNDDGEKAGVPSKR---KRSDKDGSGDDDSDDGGED 430
Score = 50.1 bits (118), Expect = 3e-06
Identities = 38/135 (28%), Positives = 55/135 (40%), Gaps = 4/135 (2%)
Frame = +2
Query: 202 DDAEDDDDDADYEEDEP----DEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDED 257
DD EDDD D D ++D P D+DD +D + E G G D + +D D+ DD +E+
Sbjct: 41 DDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEG-GRGGGGDPDDDDDDDDDDDEEE 217
Query: 258 IDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAED 317
+ D Y VR T A++ S FE E++ D
Sbjct: 218 EEEEDLGTEYL---------------VRRT------VAAAEDEEASSDFEPEEDEGDDND 334
Query: 318 SDSESDEDFKSLKKR 332
+D S +KR
Sbjct: 335 NDDGEKAGVPSKRKR 379
>TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, partial (3%)
Length = 504
Score = 48.1 bits (113), Expect = 1e-05
Identities = 37/134 (27%), Positives = 52/134 (38%)
Frame = +3
Query: 193 RVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVD 252
RV+ R +D D D EDE D+DD DD D E G G + D + D+
Sbjct: 24 RVYEIGRCRPTDDDSDPDDAESEDEEDDDDDDDGDDEDPLLGPGLGRESDSDDMSHDDY- 200
Query: 253 DSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDEN 312
DS DID D D + D F + + +++D++
Sbjct: 201 DSASDIDDEDADFILDDLD---------------------FDGGTGLLEIVGDRDEDDDD 317
Query: 313 STAEDSDSESDEDF 326
S +S S DEDF
Sbjct: 318 SQELESASSDDEDF 359
Score = 38.9 bits (89), Expect = 0.007
Identities = 34/130 (26%), Positives = 49/130 (37%)
Frame = +3
Query: 99 DDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYG 158
DD D +EDE +D+ + +DED G G+ DSD + + D +
Sbjct: 57 DDDSDPDDAESEDEEDDDDDDD------GDDEDPLLGPGLGRESDSDDMSHDDY-DSASD 215
Query: 159 QDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEP 218
D E+ + F TG + D EDDDD + E
Sbjct: 216 IDDEDADFILDDLDFDGGTGLLEI-----------------VGDRDEDDDDSQELESASS 344
Query: 219 DEDDPDDADF 228
D++D DD DF
Sbjct: 345 DDEDFDDNDF 374
Score = 35.4 bits (80), Expect = 0.077
Identities = 33/129 (25%), Positives = 53/129 (40%), Gaps = 1/129 (0%)
Frame = +3
Query: 137 GKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHR 196
G+ + D D ++ S+D D ++ G+ P G + R
Sbjct: 39 GRCRPTDDDSDPDDAESEDEEDDDDDDDGDDED-----PLLGPG-------------LGR 164
Query: 197 KSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDD-SD 255
+S D + DD D A +DE + DD DF+ T +D + +DS E++ S
Sbjct: 165 ESDSDDMSHDDYDSASDIDDEDADFILDDLDFDGGTGLLEIVGDRDEDDDDSQELESASS 344
Query: 256 EDIDVSDND 264
+D D DND
Sbjct: 345 DDEDFDDND 371
>TC12209 similar to UP|O60177 (O60177) DEAD box helicase, partial (6%)
Length = 550
Score = 44.3 bits (103), Expect = 2e-04
Identities = 21/71 (29%), Positives = 35/71 (48%)
Frame = +1
Query: 726 KIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDK 785
K W +++DEA +KN A + +TGTP+ N+V EL++L+HFL
Sbjct: 268 KSTWYRIILDEAQCIKNKSTGAARACCTLQAIYRFCLTGTPMMNNVGELYSLIHFLRIKP 447
Query: 786 FKSKDEFAQNY 796
+ F ++
Sbjct: 448 YNEWKRFQDSF 480
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 43.5 bits (101), Expect = 3e-04
Identities = 23/73 (31%), Positives = 39/73 (52%)
Frame = +3
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVS 261
++ E+DDD+ + +EDE D+DD DD D +GE+ ++ DD DE+ + S
Sbjct: 249 NEEEEDDDEEEDDEDEDDDDDDDDDD----------------DGEEEEDDDDDDEEEEGS 380
Query: 262 DNDDLYFDKKAKG 274
+ ++ AKG
Sbjct: 381 EEVEVEGKDGAKG 419
Score = 32.7 bits (73), Expect = 0.50
Identities = 14/53 (26%), Positives = 27/53 (50%)
Frame = +3
Query: 85 DCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSG 137
D P Q +E +D D +ED+ +D+ + GE+ +D+D +++ G
Sbjct: 219 DGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEG 377
Score = 29.6 bits (65), Expect = 4.2
Identities = 15/62 (24%), Positives = 28/62 (44%)
Frame = +3
Query: 88 PMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVP 147
P P ++ ++ + D +DE ED+ + + G E+ED D + + G +V
Sbjct: 216 PDGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEED-DDDDDEEEEGSEEVE 392
Query: 148 AE 149
E
Sbjct: 393 VE 398
Score = 28.5 bits (62), Expect = 9.4
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Frame = +3
Query: 109 NEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSD--DSYGQDG 161
NE+E +D+ E E +D+D D G+ + D D EE S+ + G+DG
Sbjct: 249 NEEEEDDDEEED-DEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDG 410
>TC11839 similar to UP|STH1_YEAST (P32597) Nuclear protein STH1/NPS1,
partial (4%)
Length = 519
Score = 42.7 bits (99), Expect = 5e-04
Identities = 29/78 (37%), Positives = 42/78 (53%), Gaps = 15/78 (19%)
Frame = +2
Query: 768 QNSVEELWALLHFLDSDKFKSKDEFAQNY-KNLSSFNENE--------------LSNLHM 812
+NS+EELWALL+FL + F S ++F+Q + K S +N ++ LH
Sbjct: 212 KNSLEELWALLNFLLPNIFNSSEDFSQWFNKPFESSGDNSADEALLSEEENLLIINRLHQ 391
Query: 813 ELRPHMLRRVIKDVEKSL 830
LRP +LRR+ V SL
Sbjct: 392 VLRPFVLRRLKHKVLISL 445
>BP054670
Length = 367
Score = 41.6 bits (96), Expect = 0.001
Identities = 24/58 (41%), Positives = 32/58 (54%), Gaps = 1/58 (1%)
Frame = -2
Query: 209 DDADYEEDEPD-EDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDD 265
DD+D E+DE D EDD DD+D +P G G G+ +VDD ++D D DD
Sbjct: 366 DDSDPEDDESDEEDDSDDSDVDPLL-GPGF-------GDSESDVDDRSSELDDDDGDD 217
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 40.8 bits (94), Expect = 0.002
Identities = 17/35 (48%), Positives = 23/35 (65%)
Frame = +2
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRG 236
DD +DDDDD D ++D+ D+DD DD D + S G
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSDTG 187
Score = 37.7 bits (86), Expect = 0.016
Identities = 17/44 (38%), Positives = 23/44 (51%)
Frame = +2
Query: 184 QPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDAD 227
Q T +N R+ DDDDD D ++D+ D+DD DD D
Sbjct: 17 QQQLTKINTAPSGFKRLFSPKPDDDDDDDDDDDDDDDDDDDDDD 148
Score = 35.0 bits (79), Expect = 0.10
Identities = 14/35 (40%), Positives = 22/35 (62%)
Frame = +2
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRG 236
DD +DDDDD D ++D+ D+ D D + T+ +G
Sbjct: 98 DDDDDDDDDDDDDDDDDDDGDKKQPDSDTGTALKG 202
Score = 30.0 bits (66), Expect = 3.2
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +2
Query: 244 EGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRG 278
+ +D D+ DD D+D D D+DD DKK G
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSDTG 187
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 40.4 bits (93), Expect = 0.002
Identities = 16/48 (33%), Positives = 30/48 (62%)
Frame = +2
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSD 249
D+ ED+DDD D EE++ D+DD DD + E + + +K + + + ++
Sbjct: 794 DEDEDEDDDDDEEEEDDDDDDEDDEEGEGKSKSKSGSKARPGKDQPTE 937
Score = 40.4 bits (93), Expect = 0.002
Identities = 20/44 (45%), Positives = 26/44 (58%), Gaps = 4/44 (9%)
Frame = +2
Query: 202 DDAEDDDDDADYEEDEPDEDD----PDDADFEPATSGRGANKYK 241
+D E+DD+D D +EDE D+DD DD D E G G +K K
Sbjct: 764 EDIEEDDEDGDEDEDEDDDDDEEEEDDDDDDEDDEEGEGKSKSK 895
Score = 38.1 bits (87), Expect = 0.012
Identities = 23/81 (28%), Positives = 38/81 (46%)
Frame = +2
Query: 209 DDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYF 268
+ +D+E+ E D++D D ED DE DD DE+ + D+DD
Sbjct: 749 EQSDFEDIEEDDEDGD---------------------EDEDEDDDDDEE-EEDDDDDDED 862
Query: 269 DKKAKGRQRGKFGPSVRSTRD 289
D++ +G+ + K G R +D
Sbjct: 863 DEEGEGKSKSKSGSKARPGKD 925
Score = 35.8 bits (81), Expect = 0.059
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +2
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDE 250
+D +DDDD+ + ++D+ DEDD + + SG A KD E E
Sbjct: 803 EDEDDDDDEEEEDDDDDDEDDEEGEGKSKSKSGSKARPGKDQPTERPPE 949
>TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526
At2g01100/F23H14.7 {Arabidopsis thaliana;}, partial
(33%)
Length = 902
Score = 39.7 bits (91), Expect = 0.004
Identities = 37/163 (22%), Positives = 60/163 (36%), Gaps = 12/163 (7%)
Frame = +3
Query: 206 DDDDDADYEEDEPDEDDPD------------DADFEPATSGRGANKYKDWEGEDSDEVDD 253
D D ++DY+ D+ + D+D E K K W E SD D
Sbjct: 402 DSDSESDYDSDDESKRTSRRSHRKHKKRSHYDSDHEKRKEKLSKRKTKKWSSESSDFSGD 581
Query: 254 SDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENS 313
E ++ +K K + R + S S D A S R+R+ K +
Sbjct: 582 ESES---RSEEERRRKRKQKKKLRDQVSRSDSSDSDSSADEVSKRKRQHK-----RHRVT 737
Query: 314 TAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEV 356
+ SD SDE+ ++KR R S ++ S+++
Sbjct: 738 KSSGSDFSSDEENSPVRKRSHGKHHKRHRHSESSESDLSSDDI 866
Score = 33.1 bits (74), Expect = 0.38
Identities = 26/93 (27%), Positives = 42/93 (44%)
Frame = +3
Query: 295 ASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSN 354
A +R+R++K++ ++ DSDSESD D KR R + +
Sbjct: 351 AEARERKLKAA-----KHKKRADSDSESDYDSDDESKRTSRRSHRKHKKRSHYDSDHEKR 515
Query: 355 EVRSSSRTIRKVSYVESDESEGADEGTKKSQKE 387
+ + S R +K S SD S DE +S++E
Sbjct: 516 KEKLSKRKTKKWSSESSDFS--GDESESRSEEE 608
Score = 32.3 bits (72), Expect = 0.65
Identities = 39/171 (22%), Positives = 64/171 (36%), Gaps = 12/171 (7%)
Frame = +3
Query: 100 DSKSGSDYRNEDEFEDNS--SEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSY 157
DS S SDY ++DE + S S + +K D D +K K + + + E S D
Sbjct: 402 DSDSESDYDSDDESKRTSRRSHRKHKKRSHYDSDHEKRKEKLSKRKTKKWSSE--SSDFS 575
Query: 158 GQDGEEQGESVHSRGFRP----------STGSNSCLQPTSTNVNRRVHRKSRILDDAEDD 207
G + E + E R + S S+S + +R H++ R+ +
Sbjct: 576 GDESESRSEEERRRKRKQKKKLRDQVSRSDSSDSDSSADEVSKRKRQHKRHRVTKSS--- 746
Query: 208 DDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDI 258
+D+ DE + P +K +S E D S +DI
Sbjct: 747 --GSDFSSDEEN---------SPVRKRSHGKHHKRHRHSESSESDLSSDDI 866
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 39.3 bits (90), Expect = 0.005
Identities = 29/127 (22%), Positives = 57/127 (44%), Gaps = 8/127 (6%)
Frame = +3
Query: 106 DYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQ- 164
D +E+E + + GR +++ED ++++G+ + + +V E+ ++ +D EE+
Sbjct: 354 DEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEEEEEEEVVVEDEQGNEDEDEDEEEEG 533
Query: 165 -------GESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDE 217
G V S+G +G+ S + ++R ILD + D Y E
Sbjct: 534 VVVVEVKGRKVDSKGLHSGSGTPSKVAYVIPLPDKRT--LELILDKLQKKDTYGVYAEPV 707
Query: 218 PDEDDPD 224
E+ PD
Sbjct: 708 DPEELPD 728
Score = 28.5 bits (62), Expect = 9.4
Identities = 23/87 (26%), Positives = 38/87 (43%)
Frame = +3
Query: 1065 KLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEILERAEKVE 1124
++ + +GG K +L L G EE +E+ DEE + + + E+ E
Sbjct: 258 RMRMRMMMRGGGRKKKLKLVEKLNQGVEEE-EEDEEDEEEQTPHPTRGRSRVEHAQEEDE 434
Query: 1125 EKENGGEQAHELLSAFKVANFCNDEDD 1151
E+ENG E+ E + DED+
Sbjct: 435 EEENGEEEEEEEEVVVEDEQGNEDEDE 515
>TC12505 similar to UP|BAD00698 (BAD00698) Sericin 1A, partial (5%)
Length = 569
Score = 38.5 bits (88), Expect = 0.009
Identities = 23/61 (37%), Positives = 35/61 (56%), Gaps = 4/61 (6%)
Frame = -3
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFE----PATSGRGANKYKDWEGEDSDEVDDSDED 257
D A+ DD + + EE+E +E D DD D + ATS K +D + +D +E D+ DED
Sbjct: 519 DVADVDDVNENGEEEEEEEVDDDDDDVQVLPAAATSSHPKRK-RDDDADDREEEDEDDED 343
Query: 258 I 258
+
Sbjct: 342 V 340
>BI419972
Length = 498
Score = 38.1 bits (87), Expect = 0.012
Identities = 19/39 (48%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Frame = +1
Query: 202 DDAEDDDDD--ADYEEDEPDEDDPDDADFEPATSGRGAN 238
DDA+DDDDD DY DE +E DP+D +P +G G +
Sbjct: 385 DDADDDDDDEGEDYSGDEGEEADPED---DPEANGAGGS 492
Score = 37.7 bits (86), Expect = 0.016
Identities = 18/54 (33%), Positives = 28/54 (51%)
Frame = +1
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSD 255
D +DD+D D E+D+ D+DD D+ + Y EGE++D DD +
Sbjct: 343 DSETEDDEDGDDEDDDADDDDDDEGE-----------DYSGDEGEEADPEDDPE 471
Score = 30.0 bits (66), Expect = 3.2
Identities = 12/25 (48%), Positives = 17/25 (68%)
Frame = +1
Query: 241 KDWEGEDSDEVDDSDEDIDVSDNDD 265
KD E ED ++ DD D+D D D+D+
Sbjct: 340 KDSETEDDEDGDDEDDDADDDDDDE 414
Score = 28.5 bits (62), Expect = 9.4
Identities = 16/48 (33%), Positives = 23/48 (47%)
Frame = +1
Query: 90 SPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSG 137
S E GDD +D ++DE ED S + GE+ ED+ +G
Sbjct: 346 SETEDDEDGDDEDDDADDDDDDEGEDYSGD-EGEEADPEDDPEANGAG 486
>TC9621 similar to UP|Q9SIW2 (Q9SIW2) At2g16390 protein, partial (10%)
Length = 588
Score = 37.7 bits (86), Expect = 0.016
Identities = 31/113 (27%), Positives = 46/113 (40%)
Frame = +1
Query: 1007 NPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVIQKLNAEGKL 1066
NP QA+ RA R GQ + V +YR + + S EED DH+ K K+
Sbjct: 7 NPSVTRQAIGRAFRPGQTKKVFVYRLIAADSPEED-----------DHITCFKKELISKM 153
Query: 1067 EKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEILER 1119
+F+ NE F E + E DE + +L D+ + +R
Sbjct: 154 ----------WFEWNEYCGDRAFQVETVSVNECGDEFLETPMLGEDVKALYKR 282
>TC13305 weakly similar to UP|O29528 (O29528) Cobalt transport protein
(CBIQ-1), partial (8%)
Length = 488
Score = 37.0 bits (84), Expect = 0.026
Identities = 15/53 (28%), Positives = 30/53 (56%)
Frame = +2
Query: 992 NLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILE 1044
+L A + + + +P+ + + + HRIG ++VN+ + + S+EE ILE
Sbjct: 35 DLTAASRIYLMEPYLSPEIEARMIDLVHRIGPDQIVNVVKLIMHNSIEEKILE 193
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,862,562
Number of Sequences: 28460
Number of extensions: 322902
Number of successful extensions: 2978
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 2163
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2566
length of query: 1730
length of database: 4,897,600
effective HSP length: 103
effective length of query: 1627
effective length of database: 1,966,220
effective search space: 3199039940
effective search space used: 3199039940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141111.7


