

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.8 + phase: 0
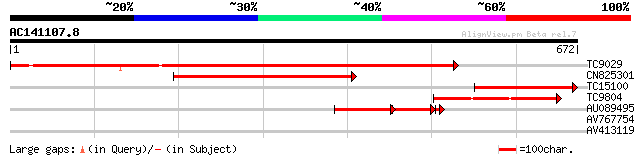
(672 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9029 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase... 936 0.0
CN825301 370 e-103
TC15100 weakly similar to UP|BAC99303 (BAC99303) Alpha-L-arabino... 195 2e-50
TC9804 similar to UP|Q84RC5 (Q84RC5) Alpha-L-arabinofuranosidase... 188 2e-48
AU089495 108 3e-35
AV767754 27 8.2
AV413119 27 8.2
>TC9029 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(73%)
Length = 1636
Score = 936 bits (2420), Expect = 0.0
Identities = 454/536 (84%), Positives = 483/536 (89%), Gaps = 4/536 (0%)
Frame = +3
Query: 1 MGFSKDSCCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFE 60
MG K S V LQL I V L ++V ADLNATL VDA + SG+ I ETLFG+FFE
Sbjct: 45 MGLYKASFIVAALQLCIAVSLCA----IKVNADLNATLTVDAHETSGKPISETLFGLFFE 212
Query: 61 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVAL 120
EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGN ++INVETDRTSCF+RNK+AL
Sbjct: 213 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIAL 392
Query: 121 RLEVLCDGT----CPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNG 176
RLEVLCD CP DGVGV+NPGFWGMNIEQGKKYKVVFY RSTG L+L VSLTGSNG
Sbjct: 393 RLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNG 572
Query: 177 VGSLASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTY 236
V LAS VITGSASDFSNW KVET+LEAKATN NSRLQLTTT KGVIWLDQVSAMPLDTY
Sbjct: 573 V--LASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTY 746
Query: 237 KGHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKY 296
KG GFRS+L+ ML DLKP FIRFPGGCFVEG++LRNAFRWK ++GPWEERPGHFGDVWKY
Sbjct: 747 KGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKY 926
Query: 297 WTDDGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPT 356
WTDDGLGY+EFLQL+EDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDG+EFARGDPT
Sbjct: 927 WTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPT 1106
Query: 357 SKWGSMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGS 416
SKWGS+RA+MGHPEPFNLKYVAVGNEDCGKKNYRGNYL+FY AIR AYPDIQIISNCDGS
Sbjct: 1107SKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGS 1286
Query: 417 SRPLDHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAAL 476
SRPLDHPAD+YDYHIYTNANDMFSRS+TFN RSGPKAFVSEYAVTGNDAG GSLLAA+
Sbjct: 1287SRPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAI 1466
Query: 477 AEAGFLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLF 532
AEAGFL+GLEKNSDIV M SYAPLFVNANDRRWNPDAIVFNSFQ YGTPSYW+QLF
Sbjct: 1467AEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLF 1634
>CN825301
Length = 663
Score = 370 bits (950), Expect = e-103
Identities = 165/217 (76%), Positives = 189/217 (87%)
Frame = +2
Query: 195 WTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGFRSDLLQMLVDLKP 254
W K+ET+LEAK TN S LQ+TT+ GV+WLDQVSAMPLDTYKGHGFR DL QM+ DL P
Sbjct: 11 WRKIETILEAKDTNHYSSLQITTSLSGVVWLDQVSAMPLDTYKGHGFRKDLFQMVADLNP 190
Query: 255 SFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDDGLGYYEFLQLSEDL 314
F RFPGGCFVEGDYLRNAFRWK VG WEERPGHFGD W YWTDDGLGY+E LQL+EDL
Sbjct: 191 RFFRFPGGCFVEGDYLRNAFRWKDTVGSWEERPGHFGDKWHYWTDDGLGYFEGLQLAEDL 370
Query: 315 GALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWGSMRAAMGHPEPFNL 374
GALP+WVFNNG+SHN+EV+T+ +LPFV EAL+G+EFARG PTS WGS RAAMGHP+PF+L
Sbjct: 371 GALPVWVFNNGISHNEEVNTTEILPFVLEALEGIEFARGSPTSYWGSRRAAMGHPKPFDL 550
Query: 375 KYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIIS 411
+ VAVGNEDC NY+GNY++FY+AI+RAYPDIQIIS
Sbjct: 551 RIVAVGNEDCSHYNYQGNYMKFYNAIKRAYPDIQIIS 661
>TC15100 weakly similar to UP|BAC99303 (BAC99303)
Alpha-L-arabinofuranosidase, partial (15%)
Length = 586
Score = 195 bits (496), Expect = 2e-50
Identities = 97/122 (79%), Positives = 110/122 (89%)
Frame = +1
Query: 551 SNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGSTKTVLTST 610
S+SL+ASAI WQ+S DKKNYIRIK VNFG + VNLKIS NGLDPNSLQ SGST+TVLTS
Sbjct: 1 SSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSA 180
Query: 611 NLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKESSNLKMLESDSSSWS 670
N+MDENSFSQPKKV+PIQSLLQ+VGK+MNV+VPPHSF+SFDLLKESSNLKM+ SS+ S
Sbjct: 181 NVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSNLKMVGGGSSTRS 360
Query: 671 SI 672
SI
Sbjct: 361 SI 366
>TC9804 similar to UP|Q84RC5 (Q84RC5) Alpha-L-arabinofuranosidase, partial
(16%)
Length = 658
Score = 188 bits (478), Expect = 2e-48
Identities = 94/152 (61%), Positives = 119/152 (77%)
Frame = +3
Query: 503 NANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQ 562
N+NDR W+PDAIVFNS Q YGTPSYW+Q F+ES+GATLL S+LQ T+S SLVASAI +
Sbjct: 3 NSNDRTWSPDAIVFNSHQHYGTPSYWLQRMFTESSGATLLKSTLQ-TSSQSLVASAIEYA 179
Query: 563 NSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPK 622
NS DKKNYIR+K VNFG+ + + +IS NGL + ++ SGS KTVLT+ N+MDENSFS+P
Sbjct: 180 NSEDKKNYIRVKVVNFGSGSESFRISINGL-KSKVKQSGSRKTVLTAANVMDENSFSEPN 356
Query: 623 KVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLK 654
K++P + ++ DM V +PPHS TSFDLLK
Sbjct: 357 KIVPQPTPIEGASTDMKVELPPHSVTSFDLLK 452
>AU089495
Length = 453
Score = 108 bits (270), Expect(3) = 3e-35
Identities = 48/73 (65%), Positives = 60/73 (81%), Gaps = 1/73 (1%)
Frame = +2
Query: 386 KKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPLDHPADMYDYHIY-TNANDMFSRSTT 444
+KNY GNYL FY+AIR+AYPDIQ+ISNCD SS+PLDHPAD+YDYH Y N ++MF+ +
Sbjct: 8 EKNYLGNYLAFYNAIRKAYPDIQMISNCDASSKPLDHPADLYDYHTYPNNPSNMFNNAHV 187
Query: 445 FNRVTRSGPKAFV 457
F++ R GPKAFV
Sbjct: 188 FDKTPRKGPKAFV 226
Score = 56.6 bits (135), Expect(3) = 3e-35
Identities = 29/52 (55%), Positives = 39/52 (74%), Gaps = 1/52 (1%)
Frame = +3
Query: 454 KAFVSEYAVTGNDAGQ-GSLLAALAEAGFLIGLEKNSDIVHMASYAPLFVNA 504
+ +SEYA+ G+ + G+L+ ++EAGFLIGL KNSD V MA+YAPLFV A
Sbjct: 216 RLLLSEYALVGDQQAKLGTLIGGVSEAGFLIGLXKNSDHVXMAAYAPLFVIA 371
Score = 21.2 bits (43), Expect(3) = 3e-35
Identities = 8/11 (72%), Positives = 10/11 (90%)
Frame = +1
Query: 505 NDRRWNPDAIV 515
+DR+ NPDAIV
Sbjct: 373 DDRKLNPDAIV 405
>AV767754
Length = 584
Score = 27.3 bits (59), Expect = 8.2
Identities = 18/62 (29%), Positives = 30/62 (48%)
Frame = -3
Query: 484 GLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLN 543
G K D+ H+A + N R + F+S ++ S+W QLF S++ G ++
Sbjct: 528 G*TKYDDVFHLACC*EMICN*YSRSIFGNDCNFHSPRMVRRSSHWTQLFSSKTLGNSVRR 349
Query: 544 SS 545
SS
Sbjct: 348 SS 343
>AV413119
Length = 391
Score = 27.3 bits (59), Expect = 8.2
Identities = 12/18 (66%), Positives = 14/18 (77%)
Frame = +1
Query: 594 PNSLQSSGSTKTVLTSTN 611
PNSL S+ ST +LTSTN
Sbjct: 46 PNSLLSTASTSPILTSTN 99
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,803,729
Number of Sequences: 28460
Number of extensions: 160531
Number of successful extensions: 722
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 717
length of query: 672
length of database: 4,897,600
effective HSP length: 96
effective length of query: 576
effective length of database: 2,165,440
effective search space: 1247293440
effective search space used: 1247293440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC141107.8


