

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.7 - phase: 0
(705 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
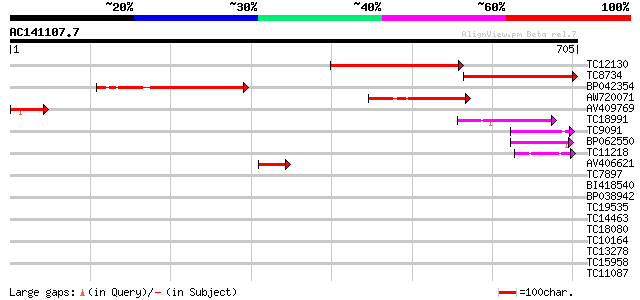
Score E
Sequences producing significant alignments: (bits) Value
TC12130 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing prot... 288 3e-78
TC8734 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing prote... 266 1e-71
BP042354 226 1e-59
AW720071 134 5e-32
AV409769 87 7e-18
TC18991 similar to UP|SUV9_ARATH (Q9T0G7) Probable histone-lysin... 74 1e-13
TC9091 similar to UP|Q9Y7R4 (Q9Y7R4) Set domain protein, transcr... 64 1e-10
BP062550 57 1e-08
TC11218 similar to UP|Q941H0 (Q941H0) Trithorax 4 (Fragment), pa... 52 2e-07
AV406621 50 2e-06
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 39 0.003
BI418540 37 0.011
BP038942 37 0.014
TC19535 similar to UP|Q80V17 (Q80V17) LOC233904 protein, partial... 36 0.018
TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein (At2g... 36 0.024
TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE... 36 0.024
TC10164 weakly similar to UP|O24099 (O24099) MtN12 protein (Frag... 35 0.041
TC13278 35 0.053
TC15958 similar to GB|AAP37838.1|30725632|BT008479 At4g24220 {Ar... 35 0.053
TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protei... 34 0.070
>TC12130 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing protein,
partial (16%)
Length = 499
Score = 288 bits (736), Expect = 3e-78
Identities = 130/165 (78%), Positives = 143/165 (85%)
Frame = +3
Query: 400 KYKLARVRGQPEAYTIWKSIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPA 459
KYKL R+ GQP+AY IWKSI QWTDK+A R GVILPDLTSGAEK+PVCLVNDVDNEKGPA
Sbjct: 3 KYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPA 182
Query: 460 YFTYIPTLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSV 519
YFTY PTLKNL +APVESS GC+C GGCQPG+ C C QKNGGYLPY+AAGL+ADLKSV
Sbjct: 183 YFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSV 362
Query: 520 IHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAI 564
++ECGPSC CPP+CRNR+SQ GLK RLE FRT KGWGLRSWD I
Sbjct: 363 VYECGPSCHCPPSCRNRVSQGGLKLRLEXFRTKGKGWGLRSWDPI 497
>TC8734 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing protein,
partial (13%)
Length = 804
Score = 266 bits (679), Expect = 1e-71
Identities = 121/141 (85%), Positives = 132/141 (92%)
Frame = +1
Query: 565 RAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYIT 624
RAGTFICEYAGEVIDNAR E L ENED+YIFDSTRIYQQLEVF +++EAPKIPSPLYIT
Sbjct: 1 RAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYIT 180
Query: 625 AKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPL 684
A+NEGNVARFMNHSC+PNVLWRP+VRENKNE DLH+AF+AIRHIPPMMELTYDYGI LPL
Sbjct: 181 ARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPL 360
Query: 685 QAGQRKKNCLCGSVKCRGYFC 705
+ GQ+KK CLCGSVKCRGYFC
Sbjct: 361 KVGQKKKKCLCGSVKCRGYFC 423
>BP042354
Length = 535
Score = 226 bits (575), Expect = 1e-59
Identities = 116/188 (61%), Positives = 146/188 (76%)
Frame = -3
Query: 109 NGDVGSSRRKSRTRRGQLTEEEGYDNTEVIDVDAETGGGSSKRKKRAKGRRASGAATDGS 168
NGDVGSSRRKSR GQL E++ + ++ +VD E G G KR+K K R ++D
Sbjct: 533 NGDVGSSRRKSR---GQLPEDDNF--VDLSEVDGEGGTGDGKRRKPQKRIREKRCSSD-- 375
Query: 169 GVAAVDVDLDAVAHDILQSINPMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQIEESTK 228
VD DAVA++IL++IN VF+++N PDGSRD+V YTLMIYEV+RRKLGQI+E K
Sbjct: 374 ------VDPDAVANEILKTINXGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAK 213
Query: 229 DLHTGAKRPDLKAGNVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHSPSMAG 288
H+GAKRPDLKAG +M TKG+R+NS+KRIG+VPGVE+GDIFFFRFE+CLVGLH+PSMAG
Sbjct: 212 GSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAG 33
Query: 289 IDYLTSKA 296
IDYL +K+
Sbjct: 32 IDYLGTKS 9
>AW720071
Length = 377
Score = 134 bits (337), Expect = 5e-32
Identities = 61/129 (47%), Positives = 87/129 (67%), Gaps = 2/129 (1%)
Frame = +3
Query: 447 CLVNDVDNEKGPAYFTYIPTLKNLRG--VAPVESSFGCSCIGGCQPGNRNCPCIQKNGGY 504
C VN +D+EK P FTYI ++ G +AP E GC C GC + CPC+ KNGG
Sbjct: 3 CAVNTIDDEKPPP-FTYITSMMYPDGCNLAPPE---GCYCTNGCSDSEK-CPCVVKNGGE 167
Query: 505 LPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAI 564
+P+ + + K +++ECGP C+CP TC NR+SQ G+KF+LE+F+T +GWG+RS ++I
Sbjct: 168 IPFNHDEAIVEAKPLVYECGPCCKCPSTCHNRVSQLGIKFQLEIFKTPTRGWGVRSLNSI 347
Query: 565 RAGTFICEY 573
+G+FICEY
Sbjct: 348 PSGSFICEY 374
>AV409769
Length = 424
Score = 87.4 bits (215), Expect = 7e-18
Identities = 43/52 (82%), Positives = 45/52 (85%), Gaps = 4/52 (7%)
Frame = +3
Query: 1 MDHNLGQESVPA----DKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPF 48
MDHNLGQ+ PA DKSRVLNVKPLRTLVPVFPSPSNPSSS+ PQGGAPF
Sbjct: 267 MDHNLGQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPF 422
>TC18991 similar to UP|SUV9_ARATH (Q9T0G7) Probable histone-lysine
N-methyltransferase, H3 lysine-9 specific 9 (Histone
H3-K9 methyltransferase 9) (H3-K9-HMTase 9) (Suppressor
of variegation 3-9 homolog 9) (Su(var)3-9 homolog 9) ,
partial (22%)
Length = 716
Score = 73.6 bits (179), Expect = 1e-13
Identities = 47/132 (35%), Positives = 72/132 (53%), Gaps = 9/132 (6%)
Frame = +3
Query: 558 LRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIF-----DSTRIYQQLEVFPANI 612
+RS D I+AG FICEY G V+ +A++L N D I+ + + L + +
Sbjct: 9 VRSLDLIQAGAFICEYTGVVLTTEQAKIL-TMNGDTLIYPGRFSERWAEWGDLSLIDSTY 185
Query: 613 EAPKIPS--PLYIT--AKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHI 668
E P PS PL NVA +++HS +PNVL + ++ ++ N H+ FA+ +I
Sbjct: 186 ERPXYPSIPPLXFAMDVSRMRNVACYISHSSTPNVLVQYVLFDHNNLMFPHLMLFAMENI 365
Query: 669 PPMMELTYDYGI 680
PPM EL+ DYG+
Sbjct: 366 PPMRELSLDYGV 401
>TC9091 similar to UP|Q9Y7R4 (Q9Y7R4) Set domain protein, transcriptional
silencing, partial (4%)
Length = 607
Score = 63.5 bits (153), Expect = 1e-10
Identities = 35/80 (43%), Positives = 46/80 (56%)
Frame = +2
Query: 623 ITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINL 682
I A GNV+RF+NHSCSP+++ ++ E+ + HI +A R I ELTYDY L
Sbjct: 2 IDASKYGNVSRFINHSCSPHLVSHQVLIESMDCERTHIGLYASRDIALGEELTYDYQYEL 181
Query: 683 PLQAGQRKKNCLCGSVKCRG 702
G CLC S+KCRG
Sbjct: 182 VPGEG---SPCLCESLKCRG 232
>BP062550
Length = 525
Score = 57.0 bits (136), Expect = 1e-08
Identities = 32/83 (38%), Positives = 42/83 (50%), Gaps = 4/83 (4%)
Frame = -3
Query: 623 ITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINL 682
I A++ GNV+RF+NHSC PN+ + ++ + + FA I P ELTYDY L
Sbjct: 439 IDAQSLGNVSRFINHSCEPNLFVQCVLSSYDDIRLARVVLFAGDDIYPYQELTYDYNYKL 260
Query: 683 PLQAGQRKK----NCLCGSVKCR 701
G KK C CG CR
Sbjct: 259 DSVIGPDKKIKQLPCHCGETTCR 191
>TC11218 similar to UP|Q941H0 (Q941H0) Trithorax 4 (Fragment), partial (26%)
Length = 535
Score = 52.4 bits (124), Expect = 2e-07
Identities = 29/76 (38%), Positives = 39/76 (51%)
Frame = +3
Query: 628 EGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAG 687
+GN+AR +NHSC PN R I+ E + I A H+ ELTYDY + +
Sbjct: 6 QGNIARLINHSCMPNCYAR-IMSLGDQEEESRIVLIAQTHVSAGEELTYDYKFEVD-ERE 179
Query: 688 QRKKNCLCGSVKCRGY 703
+ K CLC + CR Y
Sbjct: 180 EPKVPCLCKASNCRKY 227
>AV406621
Length = 126
Score = 49.7 bits (117), Expect = 2e-06
Identities = 24/41 (58%), Positives = 30/41 (72%), Gaps = 1/41 (2%)
Frame = +3
Query: 310 SGGYEDDTGDGDVLIYSGQGGVNR-EKGASDQKLERGNLAL 349
SGGYEDD +GDV+IYSG GG ++ + QKLE GNLA+
Sbjct: 3 SGGYEDDVDEGDVIIYSGHGGQDKHSRQVFHQKLEGGNLAM 125
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 38.9 bits (89), Expect = 0.003
Identities = 30/99 (30%), Positives = 44/99 (44%), Gaps = 1/99 (1%)
Frame = +3
Query: 18 LNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPF-YPFFVSPESQRL 76
++ P + P SPS+ +S++P GG + SPAG PAG P P
Sbjct: 735 VSAPPAASPAPGAESPSSSPTSNSPAGGPTATSPSPAGDSPAGGPPAPSPTTGDTPPSAS 914
Query: 77 SEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSS 115
+P+ TP + + A P +S TA G+ GSS
Sbjct: 915 GPDVSPSGTP---SSLPADTPSSSSNNSTAPPPGNGGSS 1022
>BI418540
Length = 518
Score = 37.0 bits (84), Expect = 0.011
Identities = 25/95 (26%), Positives = 37/95 (38%)
Frame = +1
Query: 28 PVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPTPQ 87
P PSP +PSS NP P + +P P + P F + P P P
Sbjct: 148 PSVPSPHSPSSLKNPSSPNPPLTPTPPPPLSLPIPPPAALFTARIGGTPIHAPPPPPAPS 327
Query: 88 RATPISAAVPINSFKTPTAATNGDVGSSRRKSRTR 122
+ AA + TP++ V + RR++ R
Sbjct: 328 PPSATPAAPHCRNPTTPSSFRTCSVNTWRRRNGMR 432
Score = 28.5 bits (62), Expect = 3.8
Identities = 25/77 (32%), Positives = 29/77 (37%)
Frame = +1
Query: 9 SVPADKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFF 68
SVP+ S P P+ P+P P S P A F A P A P P
Sbjct: 151 SVPSPHSPSSLKNPSSPNPPLTPTPPPPLSLPIPPPAALFTARIGGTPIHAPPPPPAP-- 324
Query: 69 VSPESQRLSEQHAPNPT 85
SP S + H NPT
Sbjct: 325 -SPPSATPAAPHCRNPT 372
>BP038942
Length = 617
Score = 36.6 bits (83), Expect = 0.014
Identities = 29/87 (33%), Positives = 35/87 (39%), Gaps = 1/87 (1%)
Frame = +1
Query: 20 VKPLRTLVPVFPSPSNPS-SSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPESQRLSE 78
V P +L P P S PS S SNP P +P P P +PF P RLS
Sbjct: 289 VHPSSSLTPPPPPLSPPSLSPSNPVSVLPVPLATPLSPSPP--TSLFPFLQPPTPSRLSR 462
Query: 79 QHAPNPTPQRATPISAAVPINSFKTPT 105
+ P T +S P + TPT
Sbjct: 463 LSSFTSNPSSVTSLSTK-PSSPTLTPT 540
>TC19535 similar to UP|Q80V17 (Q80V17) LOC233904 protein, partial (4%)
Length = 541
Score = 36.2 bits (82), Expect = 0.018
Identities = 20/43 (46%), Positives = 25/43 (57%)
Frame = +1
Query: 660 IAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCRG 702
I +A RHI E+TY Y PL+ ++K C CGS KCRG
Sbjct: 7 IFLYAXRHIASGEEITYXY--KFPLE--EKKIPCNCGSQKCRG 123
>TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein
(At2g16660/T24I21.7), partial (40%)
Length = 1122
Score = 35.8 bits (81), Expect = 0.024
Identities = 29/96 (30%), Positives = 42/96 (43%), Gaps = 5/96 (5%)
Frame = +2
Query: 31 PSPSNPSSSSNPQGGAPFVAV---SPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPTPQ 87
P P+N S+SS P G P A SP P P+ + P S S L P+ +
Sbjct: 134 PPPANGSASSPPSGSNPSPATTTPSPTTPTPSNPSCTSPSSSSTTSPSLKTSVKPSVSSP 313
Query: 88 RATPISAAVPINSFKTPTAATN--GDVGSSRRKSRT 121
+ P S+ + +S P+ A++ GSS S T
Sbjct: 314 ASPPTSSPLGPSSSSAPSKASSVTASSGSSLANSST 421
>TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE11_17
{Arabidopsis thaliana;}, partial (24%)
Length = 585
Score = 35.8 bits (81), Expect = 0.024
Identities = 21/66 (31%), Positives = 33/66 (49%)
Frame = -1
Query: 28 PVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPTPQ 87
P SPS+P+++ PQ + SP+ P+P+ +P P P + S Q P P P+
Sbjct: 219 PSSQSPSHPATARAPQTPSHSPPASPSSPYPSS-SPPQPHPPPPPLESPSPQPTPQPPPR 43
Query: 88 RATPIS 93
+ P S
Sbjct: 42 FSHPSS 25
>TC10164 weakly similar to UP|O24099 (O24099) MtN12 protein (Fragment),
partial (66%)
Length = 566
Score = 35.0 bits (79), Expect = 0.041
Identities = 21/74 (28%), Positives = 32/74 (42%), Gaps = 4/74 (5%)
Frame = +3
Query: 22 PLRTLVPVFPSPSNPSSSSNPQGGAPFVA----VSPAGPFPAGVAPFYPFFVSPESQRLS 77
P++T P P+P + S Q P++ +SP PFP +P P P + +
Sbjct: 33 PIQTYPPQIPNPISHSPPPPGQTYPPYIPNPIFLSPPPPFPISHSPPPPVQTYPPNIPIP 212
Query: 78 EQHAPNPTPQRATP 91
H+P P P P
Sbjct: 213 IDHSPPPVPINHAP 254
>TC13278
Length = 517
Score = 34.7 bits (78), Expect = 0.053
Identities = 21/48 (43%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Frame = +1
Query: 659 HIAFFAIRHIPPMMELTYDYGINL-----PLQAGQRKKNCLCGSVKCR 701
H AFF R I ELT+DY IN P + Q C CGS CR
Sbjct: 13 HFAFFTSRKISAQEELTWDYSINFDDHDQPAELFQ----CRCGSKFCR 144
>TC15958 similar to GB|AAP37838.1|30725632|BT008479 At4g24220 {Arabidopsis
thaliana;}, partial (52%)
Length = 800
Score = 34.7 bits (78), Expect = 0.053
Identities = 37/145 (25%), Positives = 60/145 (40%), Gaps = 21/145 (14%)
Frame = +3
Query: 31 PSPSNPSSSSNPQGGA---PFVAVSPAGPFPAGVAPFYPFFVSPESQRLSE---QHAPNP 84
PSP++ SSS++P A P ++ P P AG + P + P +++ A +P
Sbjct: 237 PSPASDSSSASPASSATASPRFSLFPTPPAAAGKSTASPAALDPPGTPITQSTTSSATSP 416
Query: 85 ---TPQRATPISAAVPINSFKT-PTAATNGDVG---------SSRRKSRTRRGQLT--EE 129
TP+ ++P S P +S PTA N + SS S TR T +
Sbjct: 417 IPTTPKPSSPSSPTSPTSSTSPGPTATPNRRIARSTAPCSETSSAPSSPTRPISAT*RSK 596
Query: 130 EGYDNTEVIDVDAETGGGSSKRKKR 154
T + + +S+R +R
Sbjct: 597 PAVSTTSALSSSSARSSSTSRRSRR 671
>TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protein
(OJ1118_G09.13 protein), partial (6%)
Length = 689
Score = 34.3 bits (77), Expect = 0.070
Identities = 29/100 (29%), Positives = 37/100 (37%)
Frame = +1
Query: 28 PVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPESQRLSEQHAPNPTPQ 87
P P+PS PSS P +P P P A SP S + AP P P
Sbjct: 70 PSTPTPSKPSSPQIPP------TSTPGSPKPPATATTN----SPSSSVSTSSGAPTPKPS 219
Query: 88 RATPISAAVPINSFKTPTAATNGDVGSSRRKSRTRRGQLT 127
R TP +P++ + +S KS T LT
Sbjct: 220 RTTP-----------SPSSNSASTTAASSSKSSTHPPSLT 306
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,253,002
Number of Sequences: 28460
Number of extensions: 188739
Number of successful extensions: 1463
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 1409
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1448
length of query: 705
length of database: 4,897,600
effective HSP length: 97
effective length of query: 608
effective length of database: 2,136,980
effective search space: 1299283840
effective search space used: 1299283840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141107.7


