

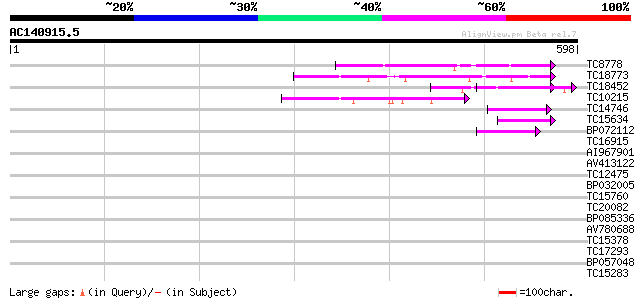
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140915.5 - phase: 0
(598 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8778 similar to UP|O22512 (O22512) Grr1, partial (31%) 64 9e-11
TC18773 similar to PIR|T48087|T48087 transport inhibitor respons... 54 6e-08
TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27... 53 1e-07
TC10215 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 48 5e-06
TC14746 weakly similar to GB|AAG21976.1|10716947|AF263377 SKP1 i... 47 7e-06
TC15634 similar to UP|Q8LGK0 (Q8LGK0) F-box protein family, AtFB... 46 2e-05
BP072112 41 6e-04
TC16915 weakly similar to UP|Q9SDA8 (Q9SDA8) AT2G17020 protein (... 35 0.035
AI967901 33 0.13
AV413122 32 0.29
TC12475 weakly similar to UP|O22512 (O22512) Grr1, partial (23%) 32 0.38
BP032005 31 0.66
TC15760 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 30 0.86
TC20082 weakly similar to UP|AAR27072 (AAR27072) EIN3-binding F-... 30 0.86
BP085336 29 1.9
AV780688 28 4.2
TC15378 similar to UP|Q941I4 (Q941I4) Vacuolar acid invertase, p... 28 4.2
TC17293 similar to GB|CAD35362.1|21535744|ATH490171 FK506 bindin... 28 4.2
BP057048 28 4.2
TC15283 similar to UP|AAR82925 (AAR82925) Coronatine-insensitive... 28 5.5
>TC8778 similar to UP|O22512 (O22512) Grr1, partial (31%)
Length = 877
Score = 63.5 bits (153), Expect = 9e-11
Identities = 67/247 (27%), Positives = 109/247 (44%), Gaps = 15/247 (6%)
Frame = +3
Query: 344 LQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQG 403
LQ FL+D+ + + + SL L C +T FF L+ NC + ++ ++ + G
Sbjct: 21 LQKCAFLSDNGLVSFAKAAPSIESLYLEECHRITQLGFFGLLFNCAAKLKV-LSLASCYG 197
Query: 404 TTIPNSLMDRLVN-PQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIR 462
N + ++ +SL + D I + L P L+ + L+ IT+ G
Sbjct: 198 IKNLNLRLPAVLPCGSIESLSIHDCPGFGDATIAVLGKLCPQLEHVELNGLSGITDAGFL 377
Query: 463 PLLES------------CRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTE-VDDEALY 509
PLLES C K+ + LSL L LEVLNL + + D +L
Sbjct: 378 PLLESSGAGLVNVNLSGCTKLT--DQVVLSLVKLHGC----SLEVLNLNGCKKISDASLI 539
Query: 510 IISNRCPALLQLVLLRCDYITDKGVMHVVN-NCTQLREINLDGCPNVQAKVVASMVVSRP 568
I+ CP L L + RC ITD G+ + + L+ ++L GC +V K V ++
Sbjct: 540 AIAGSCPLLSDLDVSRCG-ITDTGIAALAHGKQLNLKVLSLAGCTSVSNKGVRALKKLGS 716
Query: 569 SLRKIHV 575
SL +++
Sbjct: 717 SLDGLNI 737
Score = 37.7 bits (86), Expect = 0.005
Identities = 37/156 (23%), Positives = 70/156 (44%), Gaps = 2/156 (1%)
Frame = +3
Query: 313 LRRLVLSYCPGYTYSGISFLLSKSKR-IQHLDLQYTDFLNDHCVAELSLFLGDLLS-LNL 370
L + L+ G T +G LL S + +++L L D V L G L LNL
Sbjct: 324 LEHVELNGLSGITDAGFLPLLESSGAGLVNVNLSGCTKLTDQVVLSLVKLHGCSLEVLNL 503
Query: 371 GNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACL 430
C+ ++ ++ A+ +CP L++++++R I T I + +N K L LA +
Sbjct: 504 NGCKKISDASLIAIAGSCPLLSDLDVSRCGITDTGIAALAHGKQLN--LKVLSLAGCTSV 677
Query: 431 EDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLE 466
++ + L +L L++ I+ + L++
Sbjct: 678 SNKGVRALKKLGSSLDGLNIKNCKGISYRSLDMLVQ 785
>TC18773 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (50%)
Length = 927
Score = 54.3 bits (129), Expect = 6e-08
Identities = 69/301 (22%), Positives = 130/301 (42%), Gaps = 25/301 (8%)
Frame = +3
Query: 300 DQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELS 359
D L IA R LVL C G+T G++ + + + ++ LDL+ ++ + D C LS
Sbjct: 42 DDXLDLIAKSFKNFRVLVLISCEGFTTHGLAAIAANCRNLRELDLRESE-VEDICGHWLS 218
Query: 360 LFLGDLLSLNLGNCRLLT----VSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLV 415
F SL N L+ + L++ CP+L + +NR +P +DRL
Sbjct: 219 HFPDSYNSLESLNISCLSNEVNLPALERLVSRCPNLQTLRLNR------AVP---LDRLT 371
Query: 416 N-----PQFKSLFL-ASAACLEDQNIIMFAALFPNLQQLH-LSCSYNITEEGIRPLLESC 468
N PQ L A A + + + F +QL LS +++ + + C
Sbjct: 372 NLLRGAPQLVELGTGAYTAEMRPEVLANLTEAFSGCKQLKGLSGFWDVLPSYLPAVYPVC 551
Query: 469 RKIRHLNLTCLSLKS------LGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLV 522
+ LNL+ +++S + L L VL+ ++D L +++ C L +L
Sbjct: 552 SGLTSLNLSYATIQSPDHIKLVSQCGSLQRLWVLDY----IEDAGLDVLAASCKDLRELR 719
Query: 523 LLRCD--------YITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIH 574
+ D +T++G++ V C +L+ + L C + + ++ +RP++ +
Sbjct: 720 VFPSDPFGLEPNVALTEEGLISVSEGCPKLQSV-LYFCRQMSNAALNTIAQNRPNMTRFR 896
Query: 575 V 575
+
Sbjct: 897 L 899
>TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27920
{Arabidopsis thaliana;}, partial (11%)
Length = 698
Score = 53.1 bits (126), Expect = 1e-07
Identities = 38/114 (33%), Positives = 58/114 (50%), Gaps = 9/114 (7%)
Frame = +2
Query: 493 LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGC 552
LE L++T+ EVDDE L ISN C L L + C ITD+GV ++ C++L+E++L
Sbjct: 74 LEELDITDNEVDDEGLKSISN-CSRLSSLKVGICLNITDRGVAYIGMCCSKLKELDLYRS 250
Query: 553 PNVQAKVVASMVVSRPSLRKIHVPPNFPLSD---------RNRKLFSRHGCLLV 597
+ ++++ P L I+ ++D RN K GCLLV
Sbjct: 251 TGITDLGISAIACGCPGLEIINTSYCTNITDGSLISLSKCRNLKTLEIRGCLLV 412
Score = 48.5 bits (114), Expect = 3e-06
Identities = 36/138 (26%), Positives = 66/138 (47%), Gaps = 7/138 (5%)
Frame = +2
Query: 445 LQQLHLSCSYNITEEGIRPLLESCRKIRHLNL------TCLSLKSLGTNFDLPDLEVLNL 498
L L + NIT+ G+ + C K++ L+L T L + ++ P LE++N
Sbjct: 146 LSSLKVGICLNITDRGVAYIGMCCSKLKELDLYRSTGITDLGISAIACG--CPGLEIINT 319
Query: 499 TN-TEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQA 557
+ T + D +L +S +C L L + C +T G+ + NC QL +++ C N+
Sbjct: 320 SYCTNITDGSLISLS-KCRNLKTLEIRGCLLVTSIGLASIAMNCKQLICVDIKKCYNIDD 496
Query: 558 KVVASMVVSRPSLRKIHV 575
+ + SLR+I++
Sbjct: 497 SGMIPLAYFSKSLRQINL 550
Score = 28.9 bits (63), Expect = 2.5
Identities = 43/168 (25%), Positives = 66/168 (38%)
Frame = +2
Query: 94 LTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRAFSQNITTLTSLICS 153
L ++ SY D L +S LK +L + L++ + GL + + N C
Sbjct: 302 LEIINTSYCTNITDGSLISLSKCRNLK--TLEIRGCLLVTSIGLASIAMN--------CK 451
Query: 154 NLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTGLEALSLALIKLRKVNLSY 213
LI ++ + I D+ + LAY SK LR++NLSY
Sbjct: 452 QLICVDIKKCYNIDDS----GMIPLAYFSK---------------------SLRQINLSY 556
Query: 214 HGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSL 261
+ LLS +C LQ +L + L G+ ALL LT +
Sbjct: 557 SSVTDVGLLSLASISC--LQSFTILHLQGLAPGGLAAALLACRGLTKV 694
>TC10215 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (43%)
Length = 837
Score = 47.8 bits (112), Expect = 5e-06
Identities = 56/228 (24%), Positives = 98/228 (42%), Gaps = 29/228 (12%)
Frame = +3
Query: 287 GLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQY 346
GL L L +SD+ L ++ + + LVL C G+T G++ + + + ++ LDLQ
Sbjct: 51 GLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRFLRELDLQE 230
Query: 347 TDFLNDHCVAELSLF---LGDLLSLNLGNCR-LLTVSTFFALITNCPSLTEINMNRT--- 399
+ + D LS F L+SLN + + + L+ P+L + +NR+
Sbjct: 231 NE-VEDQKGQWLSCFPDSCTSLVSLNFACLKGDVNLGALERLVARSPNLKSLRLNRSVPI 407
Query: 400 -NIQ--------------GTTIPNSLMD---RLVNPQFKSLFLASAACLEDQNIIMFAAL 441
+Q G+ + N D +L N KS + S + + A+
Sbjct: 408 EALQRILMQAPQLADLGIGSFVHNPFSDAFSKLKNTILKSKSITSLSGFLEVGPFCLPAM 587
Query: 442 FP---NLQQLHLSCSYNITEEGIRPLLESCRKIRHL-NLTCLSLKSLG 485
+P NL L+LS + I + L+ C K++ L + C+ K LG
Sbjct: 588 YPICRNLTALNLSYAAGIHGNELIKLIYHCGKLQRLWIMDCIGDKGLG 731
Score = 38.9 bits (89), Expect = 0.002
Identities = 22/57 (38%), Positives = 30/57 (52%)
Frame = +3
Query: 493 LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
LE L L V DE+L ++S LVL+ C+ T G+ V NC LRE++L
Sbjct: 54 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRFLRELDL 224
>TC14746 weakly similar to GB|AAG21976.1|10716947|AF263377 SKP1 interacting
partner 1 {Arabidopsis thaliana;}, partial (62%)
Length = 719
Score = 47.4 bits (111), Expect = 7e-06
Identities = 21/67 (31%), Positives = 40/67 (59%)
Frame = +3
Query: 505 DEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMV 564
D +L +++ RCP L L + C ++TD + + NC +LRE+++ C +V K +A +
Sbjct: 384 DRSLALVAERCPNLEVLSIRSCPHVTDDSISRIAVNCPKLRELDISYCYDVTHKSLALIG 563
Query: 565 VSRPSLR 571
+ P+L+
Sbjct: 564 RNCPNLK 584
Score = 33.9 bits (76), Expect = 0.077
Identities = 18/56 (32%), Positives = 32/56 (57%), Gaps = 1/56 (1%)
Frame = +3
Query: 491 PDLEVLNLTNT-EVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLR 545
P+LEVL++ + V D+++ I+ CP L +L + C +T K + + NC L+
Sbjct: 417 PNLEVLSIRSCPHVTDDSISRIAVNCPKLRELDISYCYDVTHKSLALIGRNCPNLK 584
>TC15634 similar to UP|Q8LGK0 (Q8LGK0) F-box protein family, AtFBL4, partial
(13%)
Length = 613
Score = 45.8 bits (107), Expect = 2e-05
Identities = 20/61 (32%), Positives = 37/61 (59%)
Frame = +1
Query: 515 CPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIH 574
CP + ++V+ C ITD G+ H+V +CT L ++ C + + VA++V S +++K+
Sbjct: 1 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 180
Query: 575 V 575
V
Sbjct: 181 V 183
Score = 30.0 bits (66), Expect = 1.1
Identities = 13/43 (30%), Positives = 25/43 (57%)
Frame = +1
Query: 206 LRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGV 248
+R++ +S+ + L+HL K+C L+ ++ C +T AGV
Sbjct: 10 IREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGV 138
>BP072112
Length = 545
Score = 40.8 bits (94), Expect = 6e-04
Identities = 25/69 (36%), Positives = 32/69 (46%), Gaps = 2/69 (2%)
Frame = -1
Query: 493 LEVLNLTNT--EVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLD 550
L+V+NL D AL + C L L L CD + D GVM + C LR I+L
Sbjct: 227 LKVMNLCGCVQAASDTALQAVGQYCNQLQSLNLGWCDNVCDVGVMSLAYGCPDLRSIDLC 48
Query: 551 GCPNVQAKV 559
GC + V
Sbjct: 47 GCIRITMTV 21
Score = 30.4 bits (67), Expect = 0.86
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 1/67 (1%)
Frame = -1
Query: 508 LYIISNRCPALLQLVLLRC-DYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVS 566
L +++ C L + L C +D + V C QL+ +NL C NV V S+
Sbjct: 257 LAYLASFCRRLKVMNLCGCVQAASDTALQAVGQYCNQLQSLNLGWCDNVCDVGVMSLAYG 78
Query: 567 RPSLRKI 573
P LR I
Sbjct: 77 CPDLRSI 57
>TC16915 weakly similar to UP|Q9SDA8 (Q9SDA8) AT2G17020 protein
(At2g17020/At2g17020), partial (29%)
Length = 1011
Score = 35.0 bits (79), Expect = 0.035
Identities = 27/91 (29%), Positives = 47/91 (50%), Gaps = 3/91 (3%)
Frame = +3
Query: 480 SLKSLGTNFDLPDLEVLNLTNTEVDDEALYIIS-NRCPALLQLVLLRCDYITDKGVMHVV 538
+L+++GT L L+VL L +++ D L + + +L L L C +TDK + +
Sbjct: 99 ALQAIGT---LTKLKVLLLDGSDITDAGLSCLRPSVINSLYALSLRGCKRLTDKCIAVLF 269
Query: 539 NNC--TQLREINLDGCPNVQAKVVASMVVSR 567
+ C +LRE++L PN+ V + SR
Sbjct: 270 HGCGMLELRELDLSNLPNLSDNGVLMLAKSR 362
>AI967901
Length = 266
Score = 33.1 bits (74), Expect = 0.13
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 2/66 (3%)
Frame = +1
Query: 492 DLEVLNLTNTEV-DDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNN-CTQLREINL 549
DL LN++ D AL +++ C L L L C +H V C QL+ +NL
Sbjct: 43 DLIKLNISGCSAFSDNALAYLASFCRRLKVLNLCGCVQAASDTALHAVGQYCNQLQSLNL 222
Query: 550 DGCPNV 555
C NV
Sbjct: 223 GWCDNV 240
>AV413122
Length = 399
Score = 32.0 bits (71), Expect = 0.29
Identities = 19/78 (24%), Positives = 35/78 (44%)
Frame = +2
Query: 444 NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEV 503
+L++L L+C Y+I++EG+ + + P LE L+++ + +
Sbjct: 185 HLRRLRLACCYSISDEGLSEIAKK----------------------FPQLEELDISISSL 298
Query: 504 DDEALYIISNRCPALLQL 521
E L +I CP L L
Sbjct: 299 SKEPLEVIGRSCPRLKTL 352
>TC12475 weakly similar to UP|O22512 (O22512) Grr1, partial (23%)
Length = 501
Score = 31.6 bits (70), Expect = 0.38
Identities = 36/166 (21%), Positives = 66/166 (39%), Gaps = 7/166 (4%)
Frame = +2
Query: 362 LGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKS 421
L L SL + +CR +T A+ CP+L +++++
Sbjct: 86 LHKLKSLTIASCRGVTDVGIEAVGKGCPNLKSVHLHK----------------------- 196
Query: 422 LFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESC-RKIRHLNL-TCL 479
A L D +I F +L+ L L + IT+ G +L +C K++ +++ +C
Sbjct: 197 -----CAFLSDNGLISFTKAASSLENLQLEECHRITQFGFFGVLFNCGAKLKAISMVSCY 361
Query: 480 SLKS----LGTNFDLPDLEVLNLTN-TEVDDEALYIISNRCPALLQ 520
+K L L L++ + E + L ++ CP L Q
Sbjct: 362 GIKDQNLVLSPISPCESLRSLSIRHCPEFGNATLSVLGKLCPQLQQ 499
Score = 29.3 bits (64), Expect = 1.9
Identities = 29/139 (20%), Positives = 56/139 (39%), Gaps = 3/139 (2%)
Frame = +2
Query: 418 QFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNL- 476
+ KSL +AS + D I PNL+ +HL +++ G+ ++ + +L L
Sbjct: 92 KLKSLTIASCRGVTDVGIEAVGKGCPNLKSVHLHKCAFLSDNGLISFTKAASSLENLQLE 271
Query: 477 TCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLV-LLRCDYITDKG-V 534
C + G + + C A L+ + ++ C I D+ V
Sbjct: 272 ECHRITQFG----------------------FFGVLFNCGAKLKAISMVSCYGIKDQNLV 385
Query: 535 MHVVNNCTQLREINLDGCP 553
+ ++ C LR +++ CP
Sbjct: 386 LSPISPCESLRSLSIRHCP 442
>BP032005
Length = 449
Score = 30.8 bits (68), Expect = 0.66
Identities = 16/53 (30%), Positives = 25/53 (46%)
Frame = -2
Query: 515 CPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSR 567
CPAL +L L +C G V + C RE+ L C + V++ ++ R
Sbjct: 439 CPALERLHLQKCQLRDKNGTGAVFSVCRAAREVILQDCWGLDNSVLSLAIICR 281
>TC15760 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (17%)
Length = 664
Score = 30.4 bits (67), Expect = 0.86
Identities = 10/24 (41%), Positives = 18/24 (74%)
Frame = +2
Query: 530 TDKGVMHVVNNCTQLREINLDGCP 553
+DKG+ +V+N C +LR++ + CP
Sbjct: 77 SDKGMQYVLNGCKKLRKLEIRECP 148
>TC20082 weakly similar to UP|AAR27072 (AAR27072) EIN3-binding F-box protein
2, partial (9%)
Length = 630
Score = 30.4 bits (67), Expect = 0.86
Identities = 28/119 (23%), Positives = 54/119 (44%), Gaps = 1/119 (0%)
Frame = +3
Query: 350 LNDHCVAELSLFLGDLLSL-NLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPN 408
L D+ V+ L+ G L L NL C +T ++ A+ NC L ++++++ I T
Sbjct: 24 LTDNVVSTLARLHGGTLELLNLDGCWRITDASLVAIADNCLLLNDLDVSKCAI--TDAGL 197
Query: 409 SLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLES 467
+++ + L L+ L ++++ L L L+L +I I L+E+
Sbjct: 198 AVLSGAGQLSLQVLSLSGCCDLSNKSVAFLTKLGQTLLGLNLQHCSSIGSSTIELLVEN 374
>BP085336
Length = 369
Score = 29.3 bits (64), Expect = 1.9
Identities = 14/42 (33%), Positives = 27/42 (63%), Gaps = 1/42 (2%)
Frame = -2
Query: 444 NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSL 484
NL++LH++ +N+++ G+ L + CR + L L+ C L S+
Sbjct: 323 NLKRLHVNRCHNLSDGGLLALQDGCRSLTILYLSGCSRLTSI 198
Score = 28.5 bits (62), Expect = 3.3
Identities = 14/56 (25%), Positives = 27/56 (48%)
Frame = -2
Query: 515 CPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSL 570
C L +L + RC ++D G++ + + C L + L GC + + + RP +
Sbjct: 329 CRNLKRLHVNRCHNLSDGGLLALQDGCRSLTILYLSGCSRLTSIALELFKSHRPDV 162
>AV780688
Length = 459
Score = 28.1 bits (61), Expect = 4.2
Identities = 24/67 (35%), Positives = 34/67 (49%)
Frame = +3
Query: 19 NSSSSSELDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILY 78
NSSS+++L + ER+ R KN +HRK+ +S S+AS F C+TI
Sbjct: 12 NSSSNNDLIPKIETNERM-RTKKNLQYPNHRKKIHRSFSLASSLF-----SPDHCVTIAI 173
Query: 79 PALPVLP 85
VLP
Sbjct: 174HTDTVLP 194
>TC15378 similar to UP|Q941I4 (Q941I4) Vacuolar acid invertase, partial
(21%)
Length = 845
Score = 28.1 bits (61), Expect = 4.2
Identities = 31/102 (30%), Positives = 45/102 (43%), Gaps = 12/102 (11%)
Frame = -3
Query: 228 NCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGL---------EHLTSHF 278
NC+ L+E ++ + +A ++ L P + S+ T T L E T+
Sbjct: 354 NCQILREALMF----MPVALLNRNNLAAPYIASVG*TLPEITVLPS*AKLSMIEWSTNTL 187
Query: 279 IDSLLSLKGLTSLLLTGF---RISDQFLSSIAMESLPLRRLV 317
IDS SLK T L TGF SD +SS + + RLV
Sbjct: 186 IDSFSSLKTGTWLP*TGFATSEASDDLVSSTQKKVVEFPRLV 61
>TC17293 similar to GB|CAD35362.1|21535744|ATH490171 FK506 binding protein 1
{Arabidopsis thaliana;} , partial (21%)
Length = 575
Score = 28.1 bits (61), Expect = 4.2
Identities = 10/27 (37%), Positives = 21/27 (77%)
Frame = -2
Query: 202 ALIKLRKVNLSYHGYLNGTLLSHLFKN 228
+++K+++ NL+ HG++N TLL++ N
Sbjct: 208 SILKIKESNLAAHGFVNHTLLTNSTSN 128
>BP057048
Length = 404
Score = 28.1 bits (61), Expect = 4.2
Identities = 15/56 (26%), Positives = 31/56 (54%)
Frame = -2
Query: 14 CSQQLNSSSSSELDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNR 69
C++QL ++++ + +E FRL K + + K L++ ++ HFLS T++
Sbjct: 379 CTRQLGNNATGGTSIIVATFELSFRLFKYDSECTTSKSSLEASRGSTMHFLSKTSK 212
>TC15283 similar to UP|AAR82925 (AAR82925) Coronatine-insensitive 1, partial
(35%)
Length = 948
Score = 27.7 bits (60), Expect = 5.5
Identities = 27/120 (22%), Positives = 44/120 (36%), Gaps = 2/120 (1%)
Frame = +1
Query: 458 EEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPA 517
++G+R LL C K+R L L + D L I P
Sbjct: 190 DDGVRALLRGCDKLRRF--------------------ALYLRPGGLTDVGLGYIGQYSPN 309
Query: 518 LLQLVLLRCDYI--TDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHV 575
+ ++L Y+ TD G++ C L+++ + GC +A SLR + V
Sbjct: 310 VRWMLL---GYVGETDAGLLEFSKGCPSLQKLEIRGCSFFSEYALAIAATRLKSLRYLWV 480
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,203,550
Number of Sequences: 28460
Number of extensions: 166629
Number of successful extensions: 878
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 854
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 872
length of query: 598
length of database: 4,897,600
effective HSP length: 95
effective length of query: 503
effective length of database: 2,193,900
effective search space: 1103531700
effective search space used: 1103531700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140915.5


