

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.6 - phase: 0
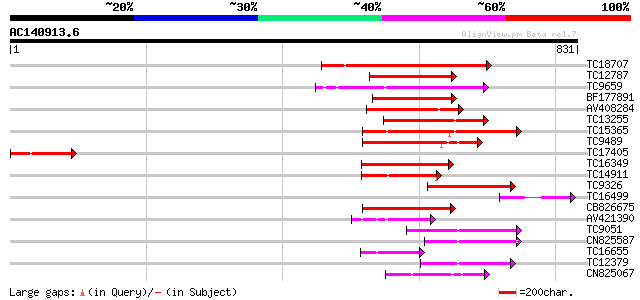
(831 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18707 homologue to PIR|T02901|T02901 MSP1 protein homolog T13J... 398 e-111
TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (T... 194 6e-50
TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATP... 174 7e-44
BF177891 169 2e-42
AV408284 169 2e-42
TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%) 158 4e-39
TC15365 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {... 156 1e-38
TC9489 homologue to UP|Q9SEI5 (Q9SEI5) 26S proteasome AAA-ATPase... 129 2e-30
TC17405 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, parti... 113 1e-25
TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulat... 112 2e-25
TC14911 homologue to UP|Q93X55 (Q93X55) Peroxin 6 (Fragment), pa... 105 3e-23
TC9326 homologue to UP|Q93WX4 (Q93WX4) Suppressor of K+ transpor... 96 2e-20
TC16499 similar to UP|Q7X9J7 (Q7X9J7) P60 katanin (Fragment), pa... 96 2e-20
CB826675 93 2e-19
AV421390 89 4e-18
TC9051 UP|PRS7_ARATH (Q9SSB5) 26S protease regulatory subunit 7 ... 82 3e-16
CN825587 79 2e-15
TC16655 homologue to UP|O99018 (O99018) Chloroplast protease pre... 78 5e-15
TC12379 homologue to UP|O04327 (O04327) Cell division protein Ft... 77 1e-14
CN825067 72 4e-13
>TC18707 homologue to PIR|T02901|T02901 MSP1 protein homolog T13J8.110 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(29%)
Length = 771
Score = 398 bits (1022), Expect = e-111
Identities = 206/249 (82%), Positives = 221/249 (88%)
Frame = +3
Query: 457 KPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGV 516
K + + ++ AP K E V K DGEN PA KAEVPDNEFEKRIRPEVIPANEIGV
Sbjct: 18 KNDVRCDNQAPENKNXTEKAIPVTKKDGENPTPA-KAEVPDNEFEKRIRPEVIPANEIGV 194
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
TF DIGA+DE K+SLQELVMLPLRRPDLF+GGLLKPCRGILLFGPPGTGKTMLA AIANE
Sbjct: 195 TFGDIGAMDEIKESLQELVMLPLRRPDLFKGGLLKPCRGILLFGPPGTGKTMLAXAIANE 374
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEA 636
AGASFINVSMSTITSKWFGEDEKNVRALF+LAAKV+PTIIFVDEVDSMLGQRTRVGEHEA
Sbjct: 375 AGASFINVSMSTITSKWFGEDEKNVRALFSLAAKVAPTIIFVDEVDSMLGQRTRVGEHEA 554
Query: 637 MRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENI 696
MRKIKNEFM++WDGL + ++ILVLAATNRPFDLDEAIIRRFERRIMVGLPS ENRE I
Sbjct: 555 MRKIKNEFMTHWDGLLTAPGEQILVLAATNRPFDLDEAIIRRFERRIMVGLPSVENREMI 734
Query: 697 LRTLLAKEK 705
L+TLLA EK
Sbjct: 735 LKTLLA*EK 761
>TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (TAT-binding
homolog 4), partial (20%)
Length = 389
Score = 194 bits (492), Expect = 6e-50
Identities = 93/128 (72%), Positives = 114/128 (88%), Gaps = 1/128 (0%)
Frame = +3
Query: 528 KDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSM 586
+D+L+ELVMLPL+RP+LF +G L KPC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+SM
Sbjct: 3 EDTLKELVMLPLKRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISM 182
Query: 587 STITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMS 646
S+ITSKWFGE EK V+A+F+LA+K+SP++IFVDEVD ML +R GEHEAMRK+ NEFM
Sbjct: 183 SSITSKWFGEGEKYVKAVFSLASKISPSVIFVDEVDXMLXRRENPGEHEAMRKMXNEFMV 362
Query: 647 NWDGLTSK 654
+WDGL SK
Sbjct: 363 HWDGLPSK 386
>TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (74%)
Length = 1060
Score = 174 bits (440), Expect = 7e-44
Identities = 101/253 (39%), Positives = 149/253 (57%)
Frame = +1
Query: 449 KKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEV 508
+ EE AV +G P PA +A + KT ++ E P+ +
Sbjct: 265 RAEEIRAVLDDG---GPGPASNGDAAVAARP-KTKPKDGGEGDGGEDPEQAKLRAGLNSA 432
Query: 509 IPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTM 568
I + + ++D+ L+ K SLQE V+LP++ P F G +P R LL+GPPGTGK+
Sbjct: 433 IIREKPNIKWNDVAGLESAKQSLQEAVILPVKFPQFFTGKR-RPWRAFLLYGPPGTGKSY 609
Query: 569 LAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR 628
LAKA+A EA ++F +VS S + SKW GE EK V LF +A + +P+IIFVDE+DS+ GQR
Sbjct: 610 LAKAVATEADSTFFSVSSSDLGSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQR 789
Query: 629 TRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLP 688
E EA R+IK E + G+ ++ ++LVLAATN P+ LD+AI RRF++RI + LP
Sbjct: 790 GEGNESEASRRIKTELLVQMQGV-GNNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLP 966
Query: 689 SAENRENILRTLL 701
+ R+++ + L
Sbjct: 967 DVKARQHMFKVHL 1005
>BF177891
Length = 390
Score = 169 bits (428), Expect = 2e-42
Identities = 82/123 (66%), Positives = 104/123 (83%), Gaps = 1/123 (0%)
Frame = +1
Query: 533 ELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITS 591
ELV+LP+RRP+LF G LL+P +GILLFGPPGTGKT++AKA+A EAGA+FI+VS ST+TS
Sbjct: 16 ELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTS 195
Query: 592 KWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGL 651
KWFG+ EK +ALF+ A+K++P IIFVDEVDS+LG R EHEA R+++NEFM+ WDGL
Sbjct: 196 KWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGL 375
Query: 652 TSK 654
SK
Sbjct: 376 RSK 384
>AV408284
Length = 425
Score = 169 bits (427), Expect = 2e-42
Identities = 86/142 (60%), Positives = 110/142 (76%), Gaps = 1/142 (0%)
Frame = +1
Query: 524 LDETKDSLQELVMLPLRRPDLFEGG-LLKPCRGILLFGPPGTGKTMLAKAIANEAGASFI 582
L+ K++L ELV+LPL+RPDLF G LL P +G+LL+GPPGTGKTMLAKAIA E+GA FI
Sbjct: 1 LESIKEALFELVILPLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFI 180
Query: 583 NVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKN 642
NV +S + SKWFG+ +K V A+F+LA K+ P IIF+DEVDS LGQR R +HEA+ +K
Sbjct: 181 NVRISNLMSKWFGDAQKLVAAVFSLAHKLQPAIIFIDEVDSFLGQR-RTTDHEALLNMKT 357
Query: 643 EFMSNWDGLTSKSEDRILVLAA 664
EFM+ WDG T+ R++VLAA
Sbjct: 358 EFMALWDGFTTDQNARVMVLAA 423
>TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%)
Length = 532
Score = 158 bits (399), Expect = 4e-39
Identities = 80/155 (51%), Positives = 110/155 (70%), Gaps = 1/155 (0%)
Frame = +1
Query: 548 GLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTL 607
GLL P +GILLFGPPGTGKTMLAKA+A E +F N+S S++ SKW G+ EK V+ LF L
Sbjct: 19 GLLSPWKGILLFGPPGTGKTMLAKAVATECKTTFFNISASSVVSKWRGDSEKLVKVLFQL 198
Query: 608 AAKVSPTIIFVDEVDSMLGQRTRV-GEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATN 666
A +P+ IF+DE+D+++ QR EHEA R++K E + DGLT ++++ + VLAATN
Sbjct: 199 ARHHAPSTIFLDEIDAIISQRGEARSEHEASRRLKTELLIQMDGLT-RTDELVFVLAATN 375
Query: 667 RPFDLDEAIIRRFERRIMVGLPSAENRENILRTLL 701
P++LD A++RR E+RI+V LP E R + LL
Sbjct: 376 LPWELDAAMLRRLEKRILVPLPEPEARVAMFEELL 480
>TC15365 homologue to GB|AAP78936.1|32306505|BT009668 At5g19990 {Arabidopsis
thaliana;}, partial (87%)
Length = 1385
Score = 156 bits (395), Expect = 1e-38
Identities = 90/240 (37%), Positives = 148/240 (61%), Gaps = 6/240 (2%)
Frame = +3
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEG-GLLKPCRGILLFGPPGTGKTMLAKAIAN 575
T+ IG LD+ ++E++ LP++ P+LFE G+ +P +G+LL+GPPGTGKT+LA+A+A+
Sbjct: 321 TYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQP-KGVLLYGPPGTGKTLLARAVAH 497
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
+FI VS S + K+ GE + VR LF +A + +P+IIF+DE+DS+ R G
Sbjct: 498 HTDCTFIRVSGSELVQKYIGEGSRMVRELFVMAREHAPSIIFMDEIDSIGSARMESGSGX 677
Query: 636 AMRKIKN---EFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
+++ E ++ DG ++ ++I VL ATNR LD+A++R R +R+I P+
Sbjct: 678 GDSEVQRTMLELLNQLDGF--EASNKIKVLMATNRIDILDQALLRPGRIDRKIEFPNPNE 851
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
E+R++IL+ + + G+D K++A G SG++LK +CT A +RE Q+D
Sbjct: 852 ESRQDILKIHSRRMNLMRGIDLKKIAEKMNGASGAELKAVCTEAGMFALRERRVHVTQED 1031
>TC9489 homologue to UP|Q9SEI5 (Q9SEI5) 26S proteasome AAA-ATPase subunit
RPT2a, partial (69%)
Length = 916
Score = 129 bits (323), Expect = 2e-30
Identities = 73/182 (40%), Positives = 113/182 (61%), Gaps = 5/182 (2%)
Frame = +1
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANE 576
+++DIG LD ++E V LPL P+L+E +KP +G++L+G PGTGKT+LAKA+AN
Sbjct: 376 SYADIGGLDAQIQEIKEAVELPLTHPELYEDIGIKPPKGVILYGEPGTGKTLLAKAVANS 555
Query: 577 AGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV---GE 633
A+F+ V S + K+ G+ K VR LF +A +SP+I+F+DE+D++ +R GE
Sbjct: 556 TSATFLRVVGSELIQKYLGDGPKLVRELFRVADDLSPSIVFIDEIDAVGTKRYDAHSGGE 735
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
E R + E ++ DG S+ + + V+ ATNR LD A++R R +R+I LP +
Sbjct: 736 REIQRTML-ELLNQLDGFDSRGD--VKVILATNRIESLDPALLRPGRIDRKIEFPLPDTK 906
Query: 692 NR 693
R
Sbjct: 907 TR 912
>TC17405 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (10%)
Length = 522
Score = 113 bits (283), Expect = 1e-25
Identities = 60/99 (60%), Positives = 80/99 (80%), Gaps = 1/99 (1%)
Frame = +1
Query: 1 MEQKGMLISA-ALSVGVGVGLGLASGQTMFKPNTYSSSSNALTPDKIENEMLRLVVDGRE 59
MEQK +L+SA ++ VG+GVGLGL+SGQ + K +S+ ++ ++I E+ +LVVDG++
Sbjct: 232 MEQKHVLLSALSVGVGLGVGLGLSSGQAVQK--WVGGNSDEVSGEQIVEEIKKLVVDGKD 405
Query: 60 SNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRN 98
S VTFD+FPYYLSE+TRVLLTSAAYVHLKH VSK+TRN
Sbjct: 406 SKVTFDDFPYYLSERTRVLLTSAAYVHLKHLNVSKHTRN 522
>TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulatory subunit
6B homolog, partial (65%)
Length = 901
Score = 112 bits (280), Expect = 2e-25
Identities = 60/137 (43%), Positives = 86/137 (61%), Gaps = 2/137 (1%)
Frame = +3
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
VT++DIG D K ++E V LPL +L++ + P RG+LL+GPPGTGKTMLAKA+AN
Sbjct: 489 VTYNDIGGCDIQKQEIREAVELPLTHHELYKQIGIDPPRGVLLYGPPGTGKTMLAKAVAN 668
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR--TRVGE 633
A+FI V S K+ GE + VR +F LA + +P IIF+DEVD++ R + G
Sbjct: 669 HTTAAFIRVVGSEFVQKYLGEGPRMVRDVFRLAKENAPAIIFIDEVDAIATARFDAQTGA 848
Query: 634 HEAMRKIKNEFMSNWDG 650
+++I E ++ DG
Sbjct: 849 DREVQRILMELLNQMDG 899
>TC14911 homologue to UP|Q93X55 (Q93X55) Peroxin 6 (Fragment), partial (32%)
Length = 1418
Score = 105 bits (262), Expect = 3e-23
Identities = 56/123 (45%), Positives = 76/123 (61%), Gaps = 6/123 (4%)
Frame = +2
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G L++ K S+ + V LPL DLF GL K G+LL+GPPGTGKT+LAKA+A
Sbjct: 215 VKWEDVGGLEDVKKSILDTVQLPLLHKDLFASGLRKRS-GVLLYGPPGTGKTLLAKAVAT 391
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDS------MLGQRT 629
E +F++V + + + GE EKNVR +F A P +IF DE+DS ML +R
Sbjct: 392 ECSLNFLSVKGPELINMYIGESEKNVRDIFQKARSARPCVIFFDELDSLAPASGMLSERV 571
Query: 630 RVG 632
VG
Sbjct: 572 LVG 580
>TC9326 homologue to UP|Q93WX4 (Q93WX4) Suppressor of K+ transport growth
defect-like protein (Fragment), partial (73%)
Length = 894
Score = 96.3 bits (238), Expect = 2e-20
Identities = 53/130 (40%), Positives = 79/130 (60%), Gaps = 1/130 (0%)
Frame = +2
Query: 613 PTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLD 672
P FVDE+DS+ GQR E EA R+IK E + G+ ++ ++LVLAATN P+ LD
Sbjct: 8 PFYHFVDEIDSLCGQRGEGNESEASRRIKTELLVQMQGV-GNNDQKVLVLAATNTPYALD 184
Query: 673 EAIIRRFERRIMVGLPSAENRENILRTLLAKEKVH-GGLDFKELATMTEGYSGSDLKNLC 731
+AI RRF++RI + LP + R+++ + L + DF+ LA+ TEG+SGSD+
Sbjct: 185 QAIRRRFDKRIYIPLPDLKARQHMFKVHLGDTPHNLNESDFEYLASRTEGFSGSDISVCV 364
Query: 732 TTAAYRPVRE 741
+ PVR+
Sbjct: 365 KDVLFEPVRK 394
>TC16499 similar to UP|Q7X9J7 (Q7X9J7) P60 katanin (Fragment), partial (61%)
Length = 646
Score = 95.9 bits (237), Expect = 2e-20
Identities = 48/112 (42%), Positives = 66/112 (58%)
Frame = +1
Query: 718 MTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERV 777
+T+GYSGSDLKNLC TAAYRPV+EL+++E D+
Sbjct: 4 LTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITN---------------------- 117
Query: 778 ITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSY 829
+ LRPLN+ DF +KSQV S A + +NELR+WN++YGEGG+R + +
Sbjct: 118 VALRPLNLDDFVQSKSQVGPSVAYDAVSVNELRKWNEMYGEGGTRTQSPFGF 273
>CB826675
Length = 551
Score = 93.2 bits (230), Expect = 2e-19
Identities = 46/138 (33%), Positives = 85/138 (61%), Gaps = 2/138 (1%)
Frame = +1
Query: 518 FSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEA 577
++D+G L++ L E ++LP+ + F+ ++P +G+LL+GPPGTGKT++A+A A +
Sbjct: 133 YNDVGGLEKQIQELVEAIVLPMTHKERFQKLGVRPPKGVLLYGPPGTGKTLMARACAAQT 312
Query: 578 GASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR--TRVGEHE 635
A+F+ ++ + + G+ K VR F LA + SP IIF+DE+D++ +R + V
Sbjct: 313 NATFLKLAGPQLVQMFIGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKRFDSEVSGDR 492
Query: 636 AMRKIKNEFMSNWDGLTS 653
+++ E ++ DG +S
Sbjct: 493 EVQRTMLELLNQLDGFSS 546
>AV421390
Length = 409
Score = 88.6 bits (218), Expect = 4e-18
Identities = 55/125 (44%), Positives = 73/125 (58%), Gaps = 2/125 (1%)
Frame = +2
Query: 502 KRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFE--GGLLKPCRGILLF 559
K + EV+P + TF D+ D+ K L+E+V LR P F GG L +GILL
Sbjct: 23 KELNKEVMPEKNVK-TFKDVKGCDDAKQELEEVVEY-LRNPAKFTRLGGKLP--KGILLT 190
Query: 560 GPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVD 619
G PGTGKT+LAKAIA EAG F + S + G + VR+LF A K +P IIF+D
Sbjct: 191 GAPGTGKTLLAKAIAGEAGVPFFYRAGSEFEEMFVGVGARRVRSLFQAAKKKAPCIIFID 370
Query: 620 EVDSM 624
E+D++
Sbjct: 371 EIDAV 385
>TC9051 UP|PRS7_ARATH (Q9SSB5) 26S protease regulatory subunit 7 (26S
proteasome subunit 7) (26S proteasome AAA-ATPase subunit
RPT1a) (Regulatory particle triple-A ATPase subunit 1a),
partial (46%)
Length = 867
Score = 82.4 bits (202), Expect = 3e-16
Identities = 54/173 (31%), Positives = 88/173 (50%), Gaps = 4/173 (2%)
Frame = +2
Query: 582 INVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR--TRVGEHEAMRK 639
I V S + K+ GE + VR LF +A I+F DEVD++ G R VG +++
Sbjct: 2 IRVIGSELVQKYVGEGARMVRELFQMARSKKACIVFFDEVDAIGGARFDDGVGGDNEVQR 181
Query: 640 IKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENRENIL 697
E ++ DG ++ I VL ATNRP LD A++R R +R++ GLP E+R I
Sbjct: 182 TMLEIVNQLDGFDARGN--IKVLMATNRPDTLDPALLRPGRLDRKVEFGLPDLESRTQIF 355
Query: 698 RTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
+ + F+ L+ + +G+D++++CT A +R + +KD
Sbjct: 356 KIHTRTMNCERDIRFELLSRLCPNSTGADIRSVCTEAGMYAIRARRKTVTEKD 514
>CN825587
Length = 663
Score = 79.3 bits (194), Expect = 2e-15
Identities = 51/147 (34%), Positives = 83/147 (55%), Gaps = 4/147 (2%)
Frame = +2
Query: 608 AAKVSPTIIFVDEVDSMLGQRTRV--GEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAAT 665
A +P I+F+DE+D++ QR G ++ + N+ ++ DG + S ++VLAAT
Sbjct: 2 AKSKAPCIVFIDEIDAVGRQRGAGLGGGNDEREQTINQLLTEMDGFSGNSG--VIVLAAT 175
Query: 666 NRPFDLDEAIIR--RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYS 723
NRP LD A++R RF+R++ V P R IL+ + + +DF ++A T G++
Sbjct: 176 NRPDVLDSALLRPGRFDRQVTVDRPDVAGRVKILQVHSRGKALAKDVDFDKIARRTPGFT 355
Query: 724 GSDLKNLCTTAAYRPVRELIQQEIQKD 750
G+DL+NL AA R + +EI KD
Sbjct: 356 GADLQNLMNEAAILAARRDL-KEISKD 433
>TC16655 homologue to UP|O99018 (O99018) Chloroplast protease precursor,
partial (39%)
Length = 934
Score = 78.2 bits (191), Expect = 5e-15
Identities = 42/94 (44%), Positives = 55/94 (57%)
Frame = +3
Query: 515 GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIA 574
GVTF D+ +DE K E+V L++P+ F + +G+LL GPPGTGKT+LAKAIA
Sbjct: 642 GVTFDDVAGVDEAKQDFMEVVEF-LKKPERFTAVGARIPKGVLLIGPPGTGKTLLAKAIA 818
Query: 575 NEAGASFINVSMSTITSKWFGEDEKNVRALFTLA 608
EAG F ++S S + G VR LF A
Sbjct: 819 GEAGVPFFSISGSEFVEMFVGVGASRVRDLFKKA 920
>TC12379 homologue to UP|O04327 (O04327) Cell division protein FtsH isolog,
partial (15%)
Length = 505
Score = 76.6 bits (187), Expect = 1e-14
Identities = 44/144 (30%), Positives = 82/144 (56%), Gaps = 4/144 (2%)
Frame = +1
Query: 602 RALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRK--IKNEFMSNWDGLTSKSEDRI 659
RAL+ A + +P+++F+DE+D++ +R + + N+ + DG + E +
Sbjct: 61 RALYQEAKENAPSVVFIDELDAVGRERGLIKGSGGQERDATLNQLLVCLDGFEGRGE--V 234
Query: 660 LVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELAT 717
+ +A+TNRP LD A++R RF+R+I + P R IL+ K+ + G +D+ +A+
Sbjct: 235 ITIASTNRPDILDPALVRPGRFDRKIFIPKPGLIGRIEILKVHARKKPIAGDVDYMAVAS 414
Query: 718 MTEGYSGSDLKNLCTTAAYRPVRE 741
MT+G G++L N+ AA +R+
Sbjct: 415 MTDGMVGAELANIVEVAAINMMRD 486
>CN825067
Length = 687
Score = 72.0 bits (175), Expect = 4e-13
Identities = 48/153 (31%), Positives = 77/153 (49%), Gaps = 1/153 (0%)
Frame = +1
Query: 552 PCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKV 611
P R +L +GPPGTGKTM A+ +A ++G + ++ + + + + LF + K
Sbjct: 187 PFRNMLFYGPPGTGKTMAARELARKSGLDYALMTGGDV-APLGSQAVTKIHELFDWSKKS 363
Query: 612 S-PTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
++F+DE D+ L +R + EA R N + +S+D +L L ATNRP D
Sbjct: 364 KRGLLLFIDEADAFLCERNKTYMSEAQRSALNALLFR---TGDQSKDIVLAL-ATNRPGD 531
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAK 703
LD A+ R + + LP R +L+ L K
Sbjct: 532 LDSAVADRIDEVLEFPLPGEGERYKLLKLYLDK 630
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,068,965
Number of Sequences: 28460
Number of extensions: 124023
Number of successful extensions: 629
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 594
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 606
length of query: 831
length of database: 4,897,600
effective HSP length: 98
effective length of query: 733
effective length of database: 2,108,520
effective search space: 1545545160
effective search space used: 1545545160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140913.6


